XG (5.25.17)
Currently Planned Experiments_5.25.17.docx
XG(5.24.17)Extra peaks XG-EB II and purposed plan.pptx
XG (5.23.17)
Question and Answer_Experimental plan_5.23.17.docx
RC (5/21): The single exp fits at beginning of your ppt "two phase exp all data points low nam mixed model"were not done correctly. What you were meant to do was to subtract the product formation at 30 mins from the product formations at all times. Thus when you subtract 30 min from the times and omitthe 0 min data point, the first data point is (0,0). I am reviewing the rest of the ppt; you can work on the scheduling below and other tasks until I finish review.
XG (5.22.17):
Experimental plan_5.22.17.docx
XG(5.19.17): Summary of the 5.18.17 meeting and updated experimental plan
Experimental plan_5.19.17.docx
Experimental plan_5.19.17_RC comments.docx
RC(5.16.17):The oaadpr, fdl high nad/NAM preliminary studies above and in xgs ppt, and the 2x e0
expt will be the exptl priorities. 2x e0 can be done 3rd. Please indicate how long each would take and amount of enzyme required for each. I will decide on dose response in the meantime.
XG(5.17.17): Experimental plan_5.17.17.docx
PMC-XG1 and 2.pptx
PMC-XG3 (1).pptx
RC (5/7): Given a) the issues with batch to batch consistency, b) the observation (described in the document below) that calculation of initial rates based on polynomial functions can significantly improve the quality of fitting to our kinetic models for this substrate,
the simulation tasks below should be postponed until after we do an additional fitting analysis to determine whether the existing data can fit our model using a polynomial function for initial rate calculation.
Recall that our previous fittings to eq 1 were all based on use of an exponential function for initial rate calculation. In the case of HKL, the quality of fitting was not very good. However, we saw based on the latest fittings posted in Dropbox that use of an exponential leads to very poor quality of fit to the (mixed) kinetic model for HKL.
1) Hence please refit -eq 1- using a second order polynomial to calculate initial rates on all the HKL data -except- the following very high NAM experiments which are not critical:
3000,8000
15000,12000 15000,15000
For this study please use -all- available time points for the 2nd order polynomial initial rate calc.
XG: Eq.1 fitting_5.7.17_wiki.pptx
2) Then, proceed with the following parts of the simulation assignment below:
--"refit the mixed model using 2nd order polynomial on 0-40 min data but omitting the expts at 100 nad / 100,200 NAM" and report the model predicted rates at the concentrations requested
--"proposal: calculate mixed model predicted rates at...report them here:"
--"using mixed model parameter estimates from 2nd order polynomial on -complete set of time series data-....report here the mixed model predicted rates..."
XG (5.9.17):Simulation result_5.9_xg.docx
Simulation 1a.b.c.pptx
RC (5/6): Please see attached for analysis of the data and reports you put in dropbox and also, the next assignment for XGResults of analysis and next assignment 5-7.docx
An edited version of the document above has been posted. In addition to some edits on pp 1-2, some edits have been made to the simulation assignment.
#1 parts 1a-c) all request model predicted rates to be posted (where it says "Report them here:").
You may post these rates before proceeding with the rest of simulation assignment #1 (which involves using cv %s to simulate noise for a-c)
RC (4/14): For XG -- addl figure task after finishing any pending ones:
eqn for 3d plot.pdf
RC: Advise on this when ready.
Also, in the double square and double triangle stacked 2d schematics, please make an alt version where k-1 is colored red and k-3 is colored blue in additionto the existing colored arrows.XG(4.20): Double square_double triangle_4.20.17.pptx
RC (4/14): This is note for XG and AU.
notes on enzyme,catalyst assay formats.docx
AU please check attachment for accuracy and add any comments/edits and any relevant refs.
AU: 4-14-2017. I have added come priliminary comments here- AU update-notes on enzyme,catalyst assay formats.docx
XG please review the edited doc and as part of your radioactivity writeup task, expand that task to include making paragraphsout of the attached bullet points (not too long, don't waste too much time, I did most of the work) and make a ref list as wellrelated to this topic.The topic is enzyme assay formats suitable for different classes of enzymes. The assays include forward and reverse reactions, and product inhibition, in continuous and discontinuous formats. The goal is to achieve higher throughput scale up of our enzyme kinetic characterization work in the future.We will use b-lactamase and sirtuin as examples as noted herein.XG(4.18): Draft out of the "Note on enzyme catalyst assay formats".
Enzyme assays overview.docx
RC(4/12):
In addition, I would like a para to be prepped on enzyme kinetics studies using radiolabeling.
It can be high level (not sirtuins only). Please add to wiki task list. In addition, we'll need some bib references for radiolabeledenzyme kinetics assays (not sirtuins only).
RC(4/10/17): Note, in the wiki task list, there was a short write up task on MST background.
XG(4.17): MST background write upMST write up.docx
Beyond this, there is a bib task for a methods paper. I am outlining some topics that need to be covered, you can post the topics on wiki and let me know when you anticipate starting it:
-MST background references
XG(4.17): Dropbox\PMC-AT References\MST background references. The references cover the following topics 1. thermophoresis theory 2. MST instrumental development 3. applications
-rapid quench methods (not specific to sirtuins, general)
XG(4.17): \Dropbox\PMC-AT References\rapid quench
-basic enzyme kinetics book
XG(4.17): \Dropbox\PMC-AT References\Enzyme kinetics textbook
-droplet technology -- RC will give some examples
You can start with the first 3 topics, assembling say 10 references from these topics
I can give more detail on some example papers that should be included once you get started.
I will also explain more about the droplet part later and give some bib entry examples. There is a related figure/schematic that will be prepped.
It is related to the plan for next steps. We are preparing a demo on such technology in May, lab staff will be involved in coordinating some of that.
XG(4/11/17):Figure modification according to comments marked in Gray below.
Double Reciprocal.Dixon Plot_4.11.17.pptx
RC(4/10/17): --Also, in the original schematic fig double reciprocal plots (parts A,C) mentioned below, an eqn for Km,S1 is shown. Can you replace that with slope = Km,S1,app/vmax <curvy equals> 1/ [E]_0 ( 1/k1,app + Kd,S1,app/k2,app ) for part A).
For part C), Km,S1,app/vmax <curvy equals> 1/ [E]_0 ( 1/k1 + Kd,S1/k2 )(1+[A]/Kd1,A / 1+[A]/Kd4,A) < Km,S1
Please see below for the equations rendered.
RC(4/10/17): Also in part D) can we make the Dixon at 15000uM NAD so we can show the exptl data? And extent x axis to same values as in part F).
Finally, remember this fig is not supposed to have 6 parts -- keep A,B part of the previous schematic fig w 4 parts and pts C-F should be A-D of a diff fig.
RC(4/10/17): We should also get an update on Thomas' prep of the real-time fig.
XG(4/10/17): SM has tried to email and call Thomas for updating. He was told that Thomas was out of town and he is back on Thursday (4/13/17). SM will follow up as soon as Thomas is back.
RC (4/7/17): Dotted lines can cross y axis unless that's cluttered. You can use judgment on that. The first Dixon in your ppt needs the eqn below next to the dotted line. The second and third Dixons need the actual numbers for plateau generated by eqn below.
RC(4/7/17): -extend the plots she made of the high NAM Dixon curves to even higher NAM so I can see the plateau more clearly
-calculate the 1/v value at plateau: this is done by using 1/v = (1/Vmax)*{ (Km/[S])* (K1/K2) + K1/K3 }
The eqn is supposed to specify horizontal line for plateau.
Please put this general form of eqn in first schematic in your ppt and in the experimental one, please substitute the actual values of the parameters as we did before to get the plateaus.
RC (4/6):
Schematic edits needed by Guan: (I am just listing these for now, they may need more clarification. By the time you finish task below, I may clarify w more details.)
--position of plateau needs to be depicted with horizontal dotted line in both original hyperbolic Dixon schematics (w A) and also in latest HKL pub quality Dixons
Part d (A=0) line of original schematics should be annotated
Annotate w eqn I provided for the plateau at sat nad
--Parts b,d (Dixons) of HKL schematics should be annotated (all lines -- 4 in total) w numbers only. Use the eqn provided w parameter estimates for HKL to get the plateau vs XG(4.7): Figure Modification_4.7.17.pptx
A brief background para on MST technique will be useful as well.
I will have some additional minor revisions to the schematic figures after the above are done.
What is status of RT MST fig?
XG(4.6.17): Sudipto has requested the raw data from 2Bind. We are waiting for Thomas' response.
RC: He needs to also ask how Thomas generated the corresponding single T jump time plot figs in the reports.
RC(4.5.17): Please do the same for any expt w two ligands --one sat and other HKL titrated. W and without other ligand compared on same axes. (MST results)
XG(4/7): Slides 4,5,6,7 are the new update.MST_Kd comparison_4.7.2017.pptx
RC(3.30.17): For every mst pair tested w wo HKL I would like
A) the fitting to be redone w both tech repeats used in single Kd fittIng
B) the HKL data pts/fitting to be plotted on the same axes as no HKL
You can post this as task on wiki.
XG(3.30.17): SM will organize the MST reports of those paired Exp.s w/w/o HKL. XG will redo the fitting A) and B).
XG(3.31.2017): Paired MST results
(1) Sirt3 + Saturating MnSOD w/w/o HKL
(2) Sirt3 + Saturating MnSOD + 1 mM NAM w/w/o HKL
(3) Sirt3 + Saturating MnSOD + 6.25uM HKL w/w/o 1mM NAM
were plotted on the same axes as requested on B).
Kd comparison_3.31.17.pptx
RC(3.30.17): Need to be able to see the intersection pt in the nonzero NAM double reciprocal plot as well.Please prepare the inset as in the other version and also change NAM to either 4000 or 5000uM so we can distinguish the lines more clearly. Ideally see both y intercepts and intersection pts of lines in inset.
Also, please make another version with 4 frames that have as a,b) the experimental plots @ 0 NAM i.e. A,B would be A,B from slide 17 and C,D would be A,B from slide 14. On B from slide 17 it seems you included some artificial points (not data).
XG (3.30.17): For double reciprocal plot ([NAM]=3000uM), the inset plot of intersection was prepared on slide 18. As RC mentioned, the intersection pt was not clear. Therefore, double reciprocal plot ([NAM]=5000uM) was prepared as well (slide19). The new version with 4 frames is prepared on Slides 16.
Figure modification_3.30.17.pptx
XG(3.30.17)
Figure modification_3.30.17.pptx
RC(3.28.17):
derepression figure task.docx
XG(3.30.17):
Depression figure.pptx
XG(3.24.2017)
Figure modification_3.24.17.pptx
XG(3.23.2017)
Plan of solubility issue_3.23.17.docx
XG(3.22.17)
Refitting_3.22.2017.pptx
RC(3.22.2017)
There are a number of methods we've been applying for which methods writeups were not prepared in the past (dls,sec,truncated,etc)
We'll need these on publication format going forward. Please arrange to prep writeups for all methods used.
XG(3.29.2017): AU and SM have prepared the draft for aforementioned methods. The write-ups have been uploaded on AU and SM's wiki page.
XG (3.21.17):
Refitting_3.21.2017_update.pptx
RC(3.20.17)
We'll also need:
--3d (like cube) figure for the triangle. ----- DONE
Here, only the mixed noncomp brackets / annotations used in the cube fig apply (no hyperbolic bracket).
Brackets would be on the left side and diagonal
Please present in same format as cube -- with the app "inset" as wellXG (3.23.17)3D Triangle Figure.pptx
--Regarding publication quality figures from exptl data mentioned,
note that HKL with this substrate displays a different behavior than the schematic reciprocal/Dixon
plots you made with annotations.
I'd like the pub quality exptl figs (reciprocal/Dixon) to be made so they can be presented alongside
the annotated schematics. Recall in this context we once had a Fig one that showed intersection pt on x-axis
in schematic form as part "a" then the experimental version as part "b" beside it.
XG(3.29.17): Done. Slide 12 in Figure modification_3.29.17.pptx on wiki/Task list from lab.
RC(3.17.2017)
Please note that the figures you made recently for Dixon based on model prediction and data points would need
to be improved to be pub quality. Of course there will be more data soon so you can decide how/when to approach this revision.
XG (3.17.17)
Figure modification_INT version.pptx
Figure modification_P2 version.pptx
RC: Please indicate which of the tasks below are addressed in these figs.
RC(3.16.2017)
In the double reciprocal and Dixon --schematics-- revision tasks I assigned:
in part b, the Dixons should curve and plateau as described in part d, unlike slide 3 in your ppt from today,
where the curves intersected with the one in presence of A/HKL starting above then plateauing below,the A curve should always lie below for all [P_1]
XG(3.17.17):Slide 11 in ppt named as "Figure modification _INT versionSlide 9 in ppt named as "Figure modification _P2 version
XG (3.15.17)
The double triangle schematic should also be prepared in color coded versions with all arrows butwith red/blue coloring analogous to that in the double square. If you have questions please let me know.
*Regarding the earlier figs sent:
-k2s not fixed to k3s in one version of square figXG (3.17.17): Done.
-Two square fig has s2p1 in middle rather than s1s2; please make other version as wellXG(3.17.17): Done.
-please make versions of all figs where INT is replaced by P2 A couple of more points regarding figs for paper.XG (3.17.17): Done. See Figure modifications_P2 version.ppt
You can post these on the wiki if needed since they aren't immediate priority but you might forget.
The priority would be the follow ups requested from XG/Alok for last Wed's high NAM ppt.
--Dixon schematics: can you redraw these by replacing the lines with the hyperbolic curves like those you plotted before for our recent analyses (see e.g. Dixon eq 1 predicted plots at 500.15000uM ppt)? You can use the equation 1 with everything except [NAD] specified and make the plot for a few diff NAD in subfigure b.
If you have any questions let me know. Later we may add depiction of location of plateau.This is not the highest priority.
XG(3.17.17):
Slide 11 in ppt named as "Figure modification _INT versionSlide 9 in ppt named as "Figure modification _P2 version
-Recall that Thomas had done some real-time MST expts for us. In these expts they get a fluorescence vs time curve like the one they show at the bottom of pg 2 in their report pdfs, but with multiple such curves in in series (one after the other). This is because they introduce multiple T jumps and measure fluorescence continuously over a longer time period.
He already gave us the raw data from such experiments, but can you ask him to send us the real-time MST plot in the style of the pg 2 bottom fig?
XG: Will do.
XG(3.13.17)
Truncated SIRT3_MM SIRTAinty_3.13.2017.pptx
XG(12.22.2016)
Updated Schedule-XG 12.22.2016.docx
XG(12.09.2016)
Updated Schedule-XG 12.09.2016.docx
XG(11.18.2016)
Updated Schedule-XG 11.18.2016.docx
XG(11.08.2016)
Updated Schedule-XG 11.08.2016.docx
XG(10.26.2016)
Updated Schedule-XG 10.26.2016.docx
NAM-HKL-SIRT3.pptx
XG(10.14.2016)
Updated Schedule-XG 10.14.2016.docx
NAM concentration optimization.pptx
XG(10.5.2016)
Updated Schedule-XG 10.04.2016.docx
Comparison of MnSOD and FdL2 peptide on Honokiol_hSIRT3.pptx
XG(9.26.2016)
PMC-XG4-2 and 3 _9.24.2016.pptx
XG(9.7.2016)
cv% PMC AU XG2.3.pptx
XG_8.23.2016
PMC-XG3 Report update.pptx
XG_8.19.2016
PMC-XG3 Report.ppt
XG_8.17.2016
PMC-XG1 and 2_Report on determination of minimum substrate concentration_8.17.16.pptx
XG_8.12.2016Report on determination of minimum substrate concentration.pptx
Schedule-XG 8.10.16.docx
XG:8.9.2016.
Schedule-XG 8.9.16_update.docx
xy value update_8.9.16_update.docx
Response to remaining questions_8.9.2016_update.docx
XG (7.29.16):Schedule update
Updated schedule_7.29.2016.docx
XG(7.28.16):xyvalue.docxUpdated schedule
Schecule updated 7.28.2016.docx
The Schedule for 3 HPLC with CRO
Things need to be considered when the experiments are designed and performed
- Since we are looking for modulator who will decrease Km NAD of SIRT3, Km measurements become important. We will compare Kms in buffer, 5% DMSO, and certain concentration of modulator in 5% DMSO.
- To minimize the systematic error, the use of same HPLC for one set of experiments is preferred. Say, we have 10 HPLCs, it will be risky to split the whole set of experiments on different HPLCs unless the calibration is done to make sure those 10 HPLC provide the same peak area for same amount sample.
- If we need to split one set samples and run on different HPLC, it will take some time to develop standard curves for data interpretation.
Initial rate experiments are the core work and time consuming.
Assuming one modulator with one concentration and do not split one set of samples for different HPLC
- 252 reactions total for Km NAD and Km peptide ---38 days (6 days for running reaction and 252/8= 31.5 days for running samples on HPLC)
- We use the second HPLC for duplicate and the third HPLC for triplicate. By doing so, the schedule will keep the same.
- Talking about CRO,
- As mentioned above, we need to make sure they calibrate their instruments well and provide solid data.
- CRO can be used for duplicate or triplicate if we do not have the third HPLC.
Assuming one modulator with one concentration and split one set of samples for different HPLCs
- 252 reactions total for Km NAD and Km peptide ---38 days (6 days for running reaction and 252/8= 31.5 days for running samples on HPLC)
- If we have CRO also with the third HPLC, 10 days will be saved.
- CRO can be used for duplicate (252 reactions)
- With repeat, there are 2X252 samples, if we have 3 HPLC, we can split them and 168 reaction per HPLC, then 27 days (6 days for running reaction and 168/8= 21 days for running samples on HPLC)
Summary of the Schedule for 2/3 HPLC w/o CRO
| Experiments |
2HPLCs |
2HPLCs + CRO |
3HPLCs |
3HPLC + CRO |
| Original reaction set (252 rnx) |
1st HPLC |
1st HPLC |
1st HPLC |
1st HPLC + 1/2 2nd HPLC |
| Duplicate (252 rnx) |
2nd HPLC |
2nd HPLC |
2nd HPLC |
3rd HPLC + 1/2 2nd HPLC |
| Triplicate (252 rnx) |
Repeat using one HPLC |
CRO |
3rd HPLC |
CRO |
| Date of Completion |
76 days |
38 days |
38 days |
27 days |
XG(7.8.16):DHP1c_In-house SIRT3 Endpoint_Repeat.pptx
RC(7.6.16): Correspondence below to be posted to wiki
FdL initial rate studies cannot be published alone. Main purpose is to compare to Hplc and understand why they are wrong/why false positives
RC (7.6.16): when are Hplc initial rates going to be done?
XG(7.7.16): The HPLC initial rates completion date is
ü 9.5.16 for one DHP1c concentration
ü 9.30.16 for two DHP1c concentrations
Please check the updated schedule listed below for details.
RC (7.6.16): why did you choose to do Hplc initial rates first (May be ok, but explain)
RC (7.7.16): Original email below should say why did you choose to do FdL initial rate expts first (not Hplc).
XG(7.7.16): There is no specific reason why FdL initial rate expts has to be done first. The experiments order can be switched.
- what is plan for duplication? When
XG(7.7.16): As soon as Sudipto have made enough enzyme, the reactions can be do parallel and will do HPLC right after the current experimental samples have finished.
- there are no dates in your schedule. Must be added
XG(7.7.16): The updated schedule with dates is listed below.
ExP.(V) ---- 7.8.16 and 7.11-7.12.16 Note: currently peptides (a-c) are available in the lab. If the 4th peptide would be shipped to lab by 7.12.16, then another day will be added for (1) and (2)
As soon as the custom synthesized peptides are delivered to lab, the use of the new FdL standard peptides in control experiments aimed at identifying why there are false positives.
(1) Background check—50uM and 250 uM peptide solution with without the addition of 1xdeveloper
(a) p53 317-320
(b) p53 317-320-Ac
(c) p53 317-320-AMC
(d) p53 317-320-Ac-AMC
(2) Standard curve using synthesized peptides and FdL standard.
Exp. (VI) Km, Vmax vs. NAD+
ü For 2 [DHP1c]s, 3x5x2x7=210 reactions (27 days) ---- 7.14.16 -- 8.19.16
ü For 1 [DHP1c], 2x5x2x7 = 140 reactions (18 days) ---- 7.14.16 – 8.6.16
[In-house SIRT3]=10ul[FdL2 peptide]=250 uM[NAD+]= 0, 100, 500, 1500, 3000uM[DHP1c]=0, 50, 100uM%DMSO=0, 5 %Time point=0, 5, 10, 20, 30, 60, 120min
Exp. (VII) Km, Vmax vs. FdL2 peptide
ü For 2 [DHP1c]s, 3x6x2x7=252 reactions (32 days) --- 8.22.16 – 9.30.16
ü For 1 [DHP1c], 2x6x2x7 = 168 reactions (21 days) ---- 8.8.16 – 9.5.16
[In-house SIRT3]=10ul[FdL2 peptide]=0, 25, 50, 100, 250, 500 uM[NAD+]= 3000uM[DHP1c]=0, 50, 100 uM% DMSO= 0, 5%Time point=0, 5, 10, 20, 30, 60, 120min
RC (7.6.16): in answer to your and aloks recent reply about p53, if it is not a sirt3 substrate, why is it used in FdL assay for sirt3?
XG (7.7.16): In Enzo kit, FdL2 peptide was used as SIRT3 peptide. In their user manual (Figure below), it was claimed that among a panel of substrates patterned on p53, p65, histone H3 and Histone H4 acetylation sites, p53-320 was the substrate deacetylated most efficiently by SIRT3. Although p53 is not a native substrate of SIRT3, it can be used for the purpose of drug discovery and screening.
RC (7.6.16): All above should be addressed on wiki. Answers will determine whether or not any if your data will be possible to publish in an upcoming submission.
RC (7.7.16): If after assessing schedule you find you cannot finish the Hplc experiments with and without dhp within about 1 month, you may consider prioritizing the Hplc over the FdL initial rate experiments. But consider the effect on aloks experimental schedule in that case as well - would it delay his work by several additional weeks.
RC:Given that you see no notable effect of DHP 1c on SIRT1 activity, and assuming that you may not see any notable effect on SIRT3 activity, you will need to consider the comments I made below before choosing a DHP 1c concentration(s) for subsequent initial rate studies. Note that EC1.5 of DHP for SIRT1 was reported to be much lower than that of SIRT3 - but still you see no effect.“-Check aloks old results to see if he saw any inhibition by DHP at those concentrations on Hplc .The only concern is that DHP may only weakly bind to sirt at those concentrations and there may be little effect.If you want to use a concentration somewhat higher than Ec1.5 for DHP that were also reported to show activation you can, in order to make sure we see some effect on initial rate parameters.”Below, you proposed to look at higher [DHP] in a dose response study, but your initial rate plan (expt V below) uses 50uM DHP.You should consider this before starting any initial rate studies with DHP/SIRT3.Also, I believe you mentioned that you would be incorporating a plan for use of the new FdL standard peptides we ordered in control experiments aimed at identifying why there are false positives.I assume you are planning to incorporate this in the plan (has not yet been done).
XG (6.29.16): Endpoint/dose response Exp. # IV is to find suitable DHP1c concentration for initial rate experiments. The results shown that in the presence of 100 and 200uM DHP1c in 5% DMSO solution, ~ 114% activation of in-house SIRT3 was detected. It’s necessary to repeat these experiments to confirm the finding. The repeat experiments will be focused on 100 and 200uM DHP1c using HPLC. The experimental conditions are listed below
REPEAT _ Exp (IV) ------- 2x2x2x3=24 reactions (3days)
[In-house SIRT3]=10U
[NAD+]=250, 2000uM
[FdL2 peptide] = 50, 250uM
% DMSO=5%
[DHP1c]=0, 100, 200uM
Time point=60min
Temp.= 37oC
ExP.(V)
As soon as the custom synthesized peptides are delivered to lab, the use of the new FdL standard peptides in control experiments aimed at identifying why there are false positives.
(1) Background check—50uM and 250 uM peptide solution with without the addition of 1xdeveloper
(a) p53 317-320
(b) p53 317-320-Ac
(c) p53 317-320-AMC
(d) p53 317-320-Ac-AMC
(2) Standard curve using synthesized peptides and FdL standard.
Exp. (VI) Km, Vmax vs. NAD+------- 3x5x2x7=210 reactions
[In-house SIRT3]=10U
[FdL2 peptide]=250 uM
[NAD+]= 0, 100, 500, 1500, 3000uM
[DHP1c]=0, 50, 100uM
%DMSO=0, 5 %
Time point=0, 5, 10, 20, 30, 60, 120min
Ex/Em= 360nm/460nm
Exp. (VII) Km, Vmax vs. FdL2 peptide------- 3x6x2x7=252 reactions
[In-house SIRT3]=10U
[FdL2 peptide]=0, 25, 50, 100, 250, 500 uM
[NAD+]= 2000uM
[DHP1c]=0, 50, 100 uM
% DMSO= 0, 5%
Time point=0, 5, 10, 20, 30, 60, 120min
Ex/Em= 360nm/460nm
RC (6.17.16): In order to run modulation assays under conditions similar to those reported for previously reported allosteric activators (resveratrol/SRT compounds), we may want to look into SI of the attached paper (which we have all seen before).
In particular, please look up the SI and determine:
a) what was [peptide] used in the EC1.5/max activation assays?
b) what was the time used in the EC1.5/max activation assays?
XG(6.17.16): SIRT1 activity was monitored using a 20 amino acid peptide (Ac-Glu-Glu-Lys(biotin)-Gly-Gln-Ser-Thr-Ser-Ser-His-Ser-Lys(Ac)-Nle-Ser-Thr-Glu-Gly–Lys(MR121 or Tamra)-Glu-Glu-NH2) derived from the sequence of p53. The mass spectrometry assay was conducted as follows: 0.5 μM peptide substrate, 120 μM βNAD+, 10 nM SIRT1, and reaction buffer (50 mM Tris-acetate pH 8, 137 mM NaCl, 2.7 mM KCl, 1 mM MgCl2, 5 mM DTT, 0.05% BSA). Reactions were incubated for 25 minutes at 25oC. Test compounds were added to the reaction or vehicle control, DMSO. After the incubation with SIRT1, 10% formic acid with 50 mM nicotinamide (Sigma) was added to stop the reaction. Milne JC et al. Nature, 45(2007)712-716.
RC(6.17.16): you also know the (p53?) peptide Km for SIRT1, please report it as well and hence the [peptide] as % of Km.
XG(6.17.16): Km value for the p53 peptide(Native peptide 3_TableS1 below) to be 4.5 uM by the HPLC method. The SIRT1 Km for NAD+ was similarly determined by HPLC and was found to be 94 uM. Data were reported in Pacholec M et al. Journal of Biological Chemistry 285(2010) 8340- 8351.
RC(6.17.16): We will use this data to compare to our assay conditions, including those for saturating peptide, nonsaturating NAD+.
XG(6.17.16): The substrate peptide used in Nature 2007 paper was fluorophore conjugated peptide. The substrate peptide used in JBC 2010 paper was native p53 peptide. The conditions used in JBC2010 paper may be used for our experiments.
*The Km of human SIRT1 enzyme for acetylated peptide substrate was examined using the SIRT1 mass spectrometry assay. To determine the Km, the linear deacetylation rate was determined at 12 concentrations of acetylated peptide substrate (50, 25, 12.5, 6.25, 3.12, 1.56, 0.78, 0.39, 0.19, 0.098, 0.049, and 0.024 μM) for each of the compound concentrations and for the vehicle control. SIRT1 enzyme, 2 mM NAD+, and 0-50 μM acetylated peptide substratewere incubated with 0-100 μM compound at 25°C. At 0, 3, 6, 9, 12, 15, 20, and 25 minutes, the reaction was stopped with 10% formic acid with 50 mM nicotinamide and the conversion of substrates to products determined by mass spectrometry. . Milne JC et al. Nature, 45(2007)712-716.
- For the Km value determinations of the native peptide, the peptide concentrations were varied from 0.78 to 100 uMand NAD+ was kept at 2 mM. For the Km value for NAD+, NAD+ was varied from 0.5 to 1000 uM and the native peptide was kept at 75 uM. Time points are 0, 5, 15,30, 45 min. The HPLC column temp. was 25oC. Pacholec M et al. Journal of Biological Chemistry 285(2010) 8340- 8351.
RC(6.17.16): Please also remind me of a) the [NAD+]'s we have used in the saturating peptide, nonsaturating NAD+ assays, along with the time (5 and 30 mins?)
XG(6.17.16): In saturating peptide, the NAD+ concentrations used were 100, 200uM for endpoint experiments at 5, 30 min time points.
b) the [peptides] we have used in nonsaturating peptide, saturating NAD+ assays.
XG(6.17.16): In saturating NAD+, the peptide concentrations used were 50uM for endpoint at 5, 30 min time points.
XG(6.15.16) updated schedule
Exp. (IV) -------------------------------- 6.13.16- 6.23.16 for experiment and 6.24.16 for report preparation
Set A (Control)-----8 reactions
[In-house SIRT3]=0, 10U
[NAD+] = 250, 2000 uM
[FdL2 peptide]= 50, 250 uM
Time point = 60 min
Temp=37oC
Set B (DMSO)------8 reactions
[In-house SIRT3]=0, 10U
[NAD+] = 250, 2000 uM
[FdL2 peptide]= 50, 250 uM
% DMSO= 5%
Time point = 60 min
Temp=37oC
Set C (DHP1c)-----40 reactions
[In-house SIRT3]=10ul
[FdL2 peptide]=50, 250 uM
[NAD+]=500, 2000 uM
[DHP1c]=0, 25, 50, 75, 100, 200uM
% DMSO= 5%
Time point=60min
Temp=37oC
Exp. (V) Km, Vmax vs. NAD+---------------6.27.16-6.28.16
[In-house SIRT3]=10ul
[FdL2 peptide]=250 uM
[NAD+]= 0, 250, 500, 1000, 2000uM
[DHP1c]=0, 50uM
%DMSO=0, 5 %
Time point=0, 5, 10, 20, 30, 60, 120min
Ex/Em= 360nm/460nm
Exp. (VI) Km, Vmax vs. FdL2 peptide-------------------6.29.16-7.1.16
[In-house SIRT3]=10ul
[FdL2 peptide]=0, 25, 50, 100, 250, 500 uM
[NAD+]= 2000uM
[DHP1c]=0, 50uM
% DMSO= 0, 5%
Time point=0, 5, 10, 20, 30, 60, 120min
Ex/Em= 360nm/460nm
Exp. (VII and VIII)Stored the sample in -80oC and run HPLC ---------------38.5 days (308reactions/7 reactions per day =38.5 days)
XG(6.10.16):
Exp. (I) ----------------------------- Done
DHP1c-SIRT1 using SIRTanity.pptx
SIRTainty assay to evaluate DHP1c/SIRT1 activation (2015 JMC)
[SIRT1]=5U
[H3K9 peptide]=25 uM
[NAD+]= 200uM
[DHP1c]=50uM
%DMSO=0, 2%
30min
Ex/Em= 420nm/460nm
Exp. (II) -------------------------------- Done
Validation of SIRTainty DHP1c.SIRT1 results by HPLC.pptx
RC: Given that you see no notable effect of DHP 1c on SIRT1 activity, and assuming that you may not see any notable effect on SIRT3 activity, you will need to consider the comments I made below before choosing a DHP 1c concentration(s) for subsequent initial rate studies. Note that EC1.5 of DHP for SIRT1 was reported to be much lower than that of SIRT3 - but still you see no effect.
“-Check aloks old results to see if he saw any inhibition by DHP at those concentrations on Hplc .The only concern is that DHP may only weakly bind to sirt at those concentrations and there may be little effect.
If you want to use a concentration somewhat higher than Ec1.5 for DHP that were also reported to show activation you can, in order to make sure we see some effect on initial rate parameters.”
Below, you proposed to look at higher [DHP] in a dose response study, but your initial rate plan (expt V below) uses 50uM DHP.
You should consider this before starting any initial rate studies with DHP/SIRT3.
Also, I believe you mentioned that you would be incorporating a plan for use of the new FdL standard peptides we ordered in control experiments aimed at identifying why there are false positives.
I assume you are planning to incorporate this in the plan (has not yet been done).
RC (6/20): To my knowledge the comments above have not yet been accounted for in the latest schedule of XG above. Please do so before proceeding with any item in the schedule that relates to the points above.
Repeat exp(C) on HPLC under same condition.
Run individual component on Beckman HPLC (old)
a. NAD+ (overlapping with exp.O-1)
b. NAM (overlapping with exp.O-1)
c. H3K9 peptide (SIRtainty kit)
d. Nicotinic acide. nicotinamidase (may not be able to run since the amount of reagent is limited)
f. Total reaction sample.
Stored the samples from Exp. (I) in -80oC and check if it’s possible to run in HPLC directly.
Reason for this experiment: To evaluate if SIRTainty data can be reproduced by HPLC. If yes, then FdL will not be used. SIRTainty assay can be used for our further use of modulator screening. If no, then we can draw conclusion that SIRTainty assay provides false-positive and DHP1c does not activate SIRT1/3.
Exp. (III) ------------------------------- Done
Comparison of deacetylation efficiency between AceCS2 and p53 by in-house SIRT3.pptx
Endpoint experiments need to be done to compare either AceCS2 or p53 deacetylated more efficiently by in-house SIRT3. Proposed conditions
[In-house SIRT3]=10 ul
[AceCS2 or p53 peptide]= 50, 600 uM
[NAD+]=100, 2000 uM
Temp.= 37oc
Time point =0, 30, 60 min
Exp. (IV) -------------------------------- 6 days
Set A (Control)-----8 reactions
[In-house SIRT3]=0, 10U
[NAD+] = 250, 2000 uM
[FdL2 peptide]= 50, 250 uM
Time point = 60 min
Temp=37oC
Set B (DMSO)------8 reactions
[In-house SIRT3]=0, 10U
[NAD+] = 250, 2000 uM
[FdL2 peptide]= 50, 250 uM
% DMSO= 5%
Time point = 60 min
Temp=37oC
Set C (DHP1c)-----20 reactions
[In-house SIRT3]=10ul
[FdL2 peptide]=50, 250 uM
[NAD+]=500, 2000 uM
[DHP1c]=0, 25, 50, 75, 100, 200uM
% DMSO= 5%
Time point=60min
Temp=37oC
Exp. (V) Km, Vmax vs. NAD+---------------2 days (5*2*2*7= 140 reactions)
[In-house SIRT3]=10ul
[FdL2 peptide]=250 uM
[NAD+]= 0, 250, 500, 1000, 2000uM
[DHP1c]=0, 50uM
%DMSO=0, 5 %
Time point=0, 5, 10, 20, 30, 60, 120min
Ex/Em= 360nm/460nm
Exp. (VI) Km, Vmax vs. FdL2 peptide-------------------3 days (6*2*2*6=168 reactions)
[In-house SIRT3]=10ul
[FdL2 peptide]=0, 25, 50, 100, 250, 500 uM
[NAD+]= 2000uM
[DHP1c]=0, 50uM
% DMSO= 0, 5%
Time point=0, 5, 10, 20, 30, 60, 120min
Ex/Em= 360nm/460nm
Exp. (VII and VIII)Stored the sample in -80oC and run HPLC ---------------38.5 days (308reactions/7 reactions per day =38.5 days)
RC (6/8): Dhp - Steegborn claimed in Ex-527 PNAS paper conclusion that they were known to be inhibitors - which paper and which DHPs? Look into the paper and check what the IC50 was, determined with which assay, it may be relevant below.
XG(6.9.16): In Ex-527 PNAS 2013 paper , it was stated “Obvious candidates, besides AGK2 and GW5074, are nicotinamide-related, Sirtuin-inhibiting 2-anilinobenzamides and dihydropyridin amides (26, 40, 41).” The reference is Mai A, et al. (2009) Study of 1,4-dihydropyridine structural scaffold: Discovery of novel sirtuin activators and inhibitors. J Med Chem 52(17):5496–5504. Figure 2 (listed below) show that
- 50uM of DHP 1a, 1b, 1d, 2a, 2b, 3a, 3b inhibit SIRT1 activity 50-85%.
- 50uM of DHP 1a, 1b, 1d inhibit SIRT2 activity ~ 50%.
- 50uM of DHP 3b and 3d inhibit SIRT3 activity ~15%
RC (6/5): To all lab staff:-As noted, need only use in house enzyme for results we plan to publish.Hence, for example, in-house enzyme not needed for all the experiments on endpoint screening of honokiol concentrations, in case the amount of available enzyme is limiting. Initial rate experiments with honokiol definitelyrequire in-house enzyme.On the other hand, we are planning to publish the repeats of DHP endpoint experiments reported in literature, and hence in-house enzyme will be required for this.
-Are Alok and Guan using different times due endpoint Hplc studies?
Lab staff planning should never reveal lack of communication (or without very good reason, lack of consensus ) in the future.AU: 6-6-2016 For endpoint assay both DHP and Honokiol, the reaction time is 30 min.
-I have given you all a relatively simple mandate on what needs to be done, in order that you may possibly generate some results that can be presented in some upcoming paper. Please see this mandate through at the least without requiring consistent monitoring, recognizing that others in the group are working on various other tasks to finish the bulk of our next publication without reliance on these results.
-All pending questions (now and in the future) will always require answers and will not be repeated.
RC (5/30):
- To all lab staff: See the PMC-AT manuscripts page for your respective tasks on figures. How many hours do you estimate it will take you to finish these? Propose a schedule of work on them. Post this by Tues am. All figures must be full done within 2 wks time max ( drafts should be provided this week).
Guan -
- Assume SIRT1/sirtainty is a false positive
- The plan to verify this through hplc is correct (endpoint). I had recommended using same conditions for HPLC and FdL from start.
- Then according to wiki schedule move on to SIRT3/DHP refutation with FdL/HPLC?
- This will include HPLC initial rate studies: when?XG(5.31.16): Yes. Right after HPLC validation for SIRTainty DHP1c/SIRT1 results.
- Will include HPLC endpoint to determine appropriate [DHP]XG(5.31.16); OK.RC: Incorporate endpoint study into schedule.
- Will also need to do control studies with fluorogenic assays to show why false positives: when will the FdL standard curves with the newly synthesized peptides be done?XG(5.31.16):When we obtain the synthesized peptides, I will provide the date.RC: You have an estimate of when they will arrive right?XG(6.1.16): We sent the order out on 5.10.16. The turnover time is 3-4weeks. The peptides are expected to be in the lab Wednesday (6.8.16).
Will there be any standard curves with sirtainty to show why false positives?XG(5.31.16): We will know if SIRTainty gave false positive after HPLC validation.
RC: - Need dates - esp when will initial rate studies, along with FdL control studies, for purpose of refutation be done?XG(6.1.16): Initial rate studies using in-house enzyme will start 6.7.16. Then when synthesized peptides come (~6.8.16) the FdL control studies can be carried on. For FdL assay initial rate studies will take 5 days. FdL control study will take 2-3 days. Then HPLC validation will take much longer time. Total 264 reactions (samples) at 7-8 samples per day, it will take days. would suggest to validate km/vmax vs. NAD+ with and without DHP1c(60 samples, 8 days for HPLC). If the HPLC results fall in agreement with FdL, then we may repeat some of the samples using HPLC upon time permission.
- HPLC resource allocation – you will not need to extensively use until we do initial rate studies with SIRT/DHP right?XG(5.31.16): Yes.
RC: On what basis do you expect FdL initial rate results to match Hplc.
You should not start any FdL initial rate assays unless you can validate with Hplc in endpoint.
Please update schedule.
XG (6.3.16): A good point. We should validate FdL results using HPLC method. The proposed endpoint experiments are listed below (The conditions are chosen from initial rate experiments, which provide duplications of some data point.)
Set A (Control)-----8 reactions
[In-house SIRT3]=0, 10U
[NAD+] = 250, 2000 uM
[FdL2 peptide]= 50, 250 uM
Time point = 60 min
Temp=37oC
Set B (DMSO)------8 reactions
[In-house SIRT3]=0, 10U
[NAD+] = 250, 2000 uM
[FdL2 peptide]= 50, 250 uM
% DMSO= 5%
Time point = 60 min
Temp=37oC
Set C (DHP1c)------8 reactions
[In-house SIRT3]=0, 10U
[NAD+] = 250, 2000 uM
[FdL2 peptide]= 50, 250 uM
[DHP1c]= 50uM
Time point = 60 min
Temp=37oC
This will take 5 days to run the reactions and HPLC.
Ideally, the amount of product formed (pmole deacetylated peptide) should be close by using (1) HPLC (% product formed calculated from peak area); (2) FdL (uM of deacetylated peptide formed calculated from standard curve). In terms of how close will be good enough, I need to look up the literatures and check if any one reported previously to claim the data from different assays fell in agreement with each other and how close they were?
After HPLC validation of endpoint experiments, we will move to initial rate experiments.
RC:Are you running FdL and HPLC on the same samples as for sirtainty below?
XG: Before I received the SIRTainty kit, I was working on running individual component (as mentioned below in Exp O1---a. NAD+; b. NAM; c. Peptide substrate (first FdL2 peptide); d. FdL standard; e. 1xDeveloper, f. AMC; g. FdL standard + 1xDeveloper) on HPLC under two concentrations to find out their retention times. I was in the middle of that. Few more components need to be run to check if there is overlapping to the product peak. Then if not, we can run FdL sample directly on HPLC.
RC: Update all dates.
XG: OK
RC:What was done this week?
XG: This week I was focusing on Validation of SIRTainty results using HPLC. The following steps were done
(1) Run individual component from SIRTainty assay
a. NAD+
b. NAM
c. H3K9 peptide (SIRtainty kit)
d. SIRTainty assay buffer
e. I did not run nicotinamidase since the amount of reagent is limited
(2) Total reaction sample from TeCan
I am now working on identifying the peaks obtained from chromatogram of total reaction sample from TeCan.
(3) I also run a set of experiments without adding SIRTainty developer to avoid the complication of the addition of developer. The samples are stored in -80oC will be run on HPLC Monday.
RC:Miscellaneous: some time ago I had asked for info on past mixed inhibition expts/plots from our work with DHP, did not receive reply (I am reiterating now since we will be redoing something like this now).
XG(6.3.16): I am looking through previous post and will response accordingly soon.
XG(5.27.16): DHP1c/SIRT1 SIRTainty experiment results is attached.DHP1c-SIRT1 using SIRTanity_5.27.2016.pptx
Schedule for next week:
üThe reactions solutions from SIRTainty have been collected and frozen into -80oC.üHPLC method need to be applied to validate the DHP1c/SIRT1 SIRTainty results. (1)Run samples collected from SIRTainty reaction (2)Prepare reactions under the same conditions for the first step. Read the finished reaction sample without adding developer.üIf the activation have been confirmed, it may suggested DHP1c is a substrate specific SIRT1 activator.üwe will move to test DHP1c/SIRT3 using H3K14 peptide (available in lab) using SIRTainty assay.
RC (5/23): It appears you have largely omitted FdL studies from the plan with DHP.So what happened to all the FdL studies you mentioned below and the standards you bought?There is no commentary on how you might provide some insight into why FdL gives false positives with DHP based eg on your control studies. Do you now plan to provide this commentary only for sirtainty? You are planning to use only sirtainty, even though sirtainty was never used with SIRT3 in the publications?
XG (5.24.16): I misunderstood your points. I have revised the related experimental design/schedule below.
Since we are refuting DHP1c activation, we should use the same peptided substrate (FdL2 peptide) and assay system (Enzo FdL assay) for comparison to HPLC results.
Sirtainty might be better than FdL for non-autofluorescent compounds, but what is the point of using it here?
XG (5.24.16): No.
Is your hope that sirtainty will not provide the false positive with SIRT3? Or, are you proposing to use sirtainty because it uses unlabeled peptide? Because it is more reproducible?
XG (5.24.16): The main reason of using SIRTainty, is to test if we can repeat 2015 JMC paper. In that paper, different peptide substrate was used. Then we may get an idea, if the DHP1c/SIRT1 activation is substrate specific.
Do you intend to combine your old FdL results with the new sirtainty results for publication? If so, are you satisfied with your old control studies for publication of FdL results?
XG (5.24.16): If the same sample from FdL can be run on HPLC, which will be strong evidence, FdL produce false positive results. SIRTainty data will be used for (1) if we see activation using SIRTanty and HPLC, then DHP1c/SIRT1 activation is substrate specific. (2) if we see activation using SIRTanty but not HPLC, then SIRTainty assay is not good for DHPs compound screening, same as FdL.
The point is to refute the published results for DHP/SIRT3. If you use other substrates (e.g. to show generality of results) or assays, it would need to be clear why and would ideally be in addition to the ones published.
XG(5.24.16): I agree.
You mentioned that the reaction may not be driven by H3K9 for SIRT3. Did you choose it rather than p53 because it was used in the JMC2015 paper (verify), though not with SIRT3?
XG(5.24.16): H3K9 was used in JMC 2015 paper for SIRT1. H3K14 was good with SIRT3 from SIRTainty
In initial rate studies, a higher concentration of DHP than EC1.5 may be used for the reasons noted below. You did not account for or mention that under Alok's old results.
XG(5.24.16):
Alok’s previous results on DHP1c/Enzo SIRT3
[NAD+]=500uM
[FdL2 peptide] =250uM
[SIRT3]=5U
5% DMSO
37oC
60min
% product formed in 0, 10, 25, 50 uM DHP1c are 2.12%, 2.06%, 1.84%, and 2.11%.
It also appears there are no initial rate studies with fluorogenic assay. This might be ok, but commentary must be provided on why and the HPLC conditions would need to directly match those with fluorogenic in at least the endpoint assays.
XG (5.24.16): If the same sample from FdL can be run on HPLC, we will do both to get direct comparable data.
You did not highlight the directly conditions, although you previously planned to run HPLC and FdL on the same samples, as I recall.
XG (5.24.16): Please see the updated schedule.Finally, there is no indication of the time required. I would need to know when each publishable result will be obtained.
XG (5.24.16): The time has been added. All the above points should be addressed carefully with an eye toward publication.I may not have time to provide further input at this point, esp since there is little commentary on the reasoning within the context of the stated goals. Please think carefully before doing experiments that take a month and result in nothing publishable. This cannot be sustained.
RC (5/22): --Following are some experimental prioritiesof our current work: -All published results must use in house enzyme.
1.
refuting honokiol activation for some substrates(not necc all; further mech studies as arranted) -With respect to your work with FdL, since Honokiol was not reported with FdL we would not need to compareto FdL results to refute. - Alok will be doing initial rate studies with p53or mnsod peptide first without honokiol then with-Since your FdL results with honokiol werenot always consistent with Hplc, we won't use FdL for publication of initial rate assays unless we see consistent effects in future with new substrates. This is ok sincewe primarily need to do two sets of initial rate exptsfor honokiol - with, without honokiol - and this can be handled on Hplc.XG(5.24.16): Alok is working on this task.
2.
refuting dhp activation -However with dhp we should compare Hplc to FdLunder the same conditions to refute. This is related to yourproposed expts.
XG(5.24.16): Ideally, the direct comparison of HPLC and FdL results can be achieved by running SAME sample on TeCan and HPLC. The proposed experiments Exp. O
Exp. (O-1)
Run individual component on Beckman HPLC (old) -----3 days (6*2+8=20 samples)
a. NAD+; b. NAM; c. Peptide substrate (first FdL2 peptide); d. FdL standard; e. 1xDeveloper, f. AMC; g. FdL standard + 1xDeveloper
Note: (1) Except FdL standard, the components will be run under 2 concentrations. 45 mins per sample + column wash + solution preparing purge the pump to get rid of bubbles, maximum 7 samples can be run for one day. (2) 8 FdL standard concentrations are needed for standard curve and detection limit of old HPLC (assuming the FdL standard is similar to product deacetylated peptide.
Exp. (O-2)
If the a-f peaks are not overlapping, then run same sample from FdL assay directly. ----- 2 days
Samples will be selected from FdL reactions (a) low NAD+ and FdL2 peptide concentrations; (b) early time point such as 5 min and 10min; (c)addition of DMSO; (d) addition of DHP1c.
-Any initial rate expts with FdL substrate will onlybe usable on context of dhp refutation. For dhpone may compare the FdL substrate with FdL and Hplcassays.
XG (5.24.16): Run initial rate experiment for in-house SIRT3 using FdL assay with and without DHP1c. 50uM DHP1c (reported EC1.5) will be used. 0 and 5% DMSO will be included.
(A-1) Km, Vmax vs. NAD+---------------2 days (5*2*2*7= 140 reactions)
[In-house SIRT3]=10ul
[FdL2 peptide]=250 uM
[NAD+]= 0, 250, 500, 1000, 2000uM
[DHP1c]=0, 50uM
%DMSO=0, 5% DMSO
0, 5, 10, 20, 30, 60, 120min
Ex/Em= 360nm/460nm
(A-2) Km, Vmax vs. FdL2 peptide--------------------3 days (6*2*2*6=168 reactions)
[In-house SIRT3]=10ul
[FdL2 peptide]=0, 25, 50, 100, 250, 500 uM
[NAD+]= 2000uM
[DHP1c]=0, 50uM
% DMSO= 0, 5%
0, 5, 10, 20, 30, 60, 120min
Ex/Em= 360nm/460nm
(B-1 and B-2)Stored the sample in -80oC and run HPLC ---------------38.5 days (308reactions/7 reactions per day =38.5 days)
-You should assume that your sirtaintyexpt with dhp will be consistent with publication but inconsistent with Hplc.Will need to be repeated w Hplc for refutation.
XG (5.24.16):
Exp. (C) ----------------------------- 2 days
SIRTainty assay to evaluate DHP1c/SIRT1 activation (2015 JMC)
[SIRT1]=5U
[H3K9 peptide]=25 uM
[NAD+]= 200uM
[DHP1c]=50uM
%DMSO=0, 5%
30min
Ex/Em= 420nm/460nm
Reason for this experiment: To evaluate if DHP1c active SIRT1 using SIRTainty assay. If the activation is spotted, then repeat Exp(C) on HPLC. If the activation is not spotted, then SIRTainty assay will not be used for modulator screening. Exp. (D) will not be performed.
Note: (1) H3K9 peptide is included in the kit, H3K14 is the one we synthesized and available in the lab. (2) This SIRTainty assay can to be run first to confirm if DHP1c activates SIRT1 using another assay system in which no fluorophore tagged peptide was used.
Exp. (D) -------------------------------- 3 days
Repeat exp(C) on HPLC under same condition.
Run individual component on Beckman HPLC (old)a. NAD+ (overlapping with exp.O-1)b. NAM (overlapping with exp.O-1)
c. H3K9 peptide (SIRtainty kit)
d. Nicotinic acid
e. nicotinamidase (may not be able to run since the amount of reagent is limited)
f. Total reaction sample
Stored the samples from Exp. (C) in -80oC and check if it’s possible to run in HPLC directly.
Reason for this experiment: To evaluate if SIRTainty data can be reproduced by HPLC. If yes, then FdL will not be used. SIRTainty assay can be used for our further use of modulator screening. If no, then we can draw conclusion that SIRTainty assay provides false-positive and DHP1c does not activate SIRT1/3.
-For purpose of publication, the priority here would beto compare dhp initial rate studies with Hplc and FdLor sirtainty and based on your more rigorous analysis of controls/individual components/ standard curves provide some intuition as to why the Fluorogenic assays provided false positive results. However you should notaim to solve this problem, since the main goalis refutation along with some explanation that increases reader confidence that we are right.
XG(5.24.16): OK. Since we refuting DHP1c activation, we will use the same peptided substrate (FdL2 peptide) and assay system (Enzo FdL assay) for comparison to HPLC results.
-In addition to refutation we may get some info onthe mode of inhibition of sirt by dhp.
XG(5.24.16): Yes. From Exp A and B we will get some info on how km, vmax, kcat change with and without DHP1c.
-You should use at least the Ec1.5 used in the papers.
XG(5.24.16): The reported EC1.5 (50uM) or number from Exp. (E) will be used in the paper.
-Check aloks old results to see if he saw any inhibitionby dhp at those concentrations on Hplc .The only concern is that dhp may only weakly bind to sirt at those concentrations and there may be little effect.
Alok’s previous results on DHP1c/Enzo SIRT3
[NAD+]=500uM
[FdL2 peptide] =250uM
[SIRT3]=5U
5% DMSO
37oC
60min
% product formed in 0, 10, 25, 50 uM DHP1c are 2.12%, 2.06%, 1.84%, and 2.11%.
If you want to use a concentration somewhat higher thanEc1.5 for dhp that were also reported to show activation you can, in order to make sure we see some effect on initial rate parameters.
XG(5.24.16): To make sure we see some effect on initial rate parameters with/without DHP1c, the concentration of DHP1c need to be modified. Because of the strong intrinsic fluorescence DHP1c has, we will run a set of dose response experiment using HPLC.
ExP. (E): ---------------------------------------- 2 days
[In-house SIRT3]=10ul
[FdL2 peptide]=250 uM
[NAD+]= 2000 uM
[DHP1c]=0, 25, 50, 75, 100, 200uM
% DMSO= 5%
60min
Ex/Em= 360nm/460nm
3.
establishing a higher throughput assay with reliableresults compared to Hplc, in absence of autofluorescence 4. for DMSO, the only remaining relevant study wouldbe MM initial rate study on Hplc to see its effect on MMparameters
XG (5.24.16): Expts. (A-1) and (A-2) will provide the results.
-Regarding your FdL/Hplc comparison you should assume you will get consistent results in absenceof DMSO and autofluorescence but the real challenge willbe in presence of DMSO since many drug screening effortswill need to use it. Demonstrating this will be usefulfor scaling up future work but not an immediate priority forpublication. -As part of the initial rate studies you should also run DMSO without dhp to show the effect of DMSO oninitial rate parameters. XG (5.24.16): Expts. (A-1) and (A-2) will provide the results.
For the purposes of our papers, 1,2,4 are higher priorities in that order.
5.
What is the plan in future due initial rate characterization of new enzyme batches - which assay and substrate?XG (5.23.16): For new batch of enzyme purified in house (scale up using FPLC), at least for the first three batches we need to test km, Vmax, Kcat. If the numbers are close enough, then we can random test for the further batches. If the numbers are not consistent, then (1) purification method modification is needed (2) keep test the newly purified batches till it’s reproducible.
The test method will be HPLC or reliable assay (SIRTainty or some assay we established in lab). The choice of the substrate will be one of the three peptides (p53, H3, and AceCS2) currently available in the lab. We need to find out which one has the best efficiency for deacetylation reaction with in-house SIRT3. When the proposed experiments have completed, we will get good idea how the assay set up.
-If you encounter any issues with reproducibility of FdL or sirtainty assays that interfere with the above plan you should immediately move to help alok finish his Hplc work with honokiol more quickly.XG(5.23.16): OK.-Please edit your schedule accordingly showing when (approx what dates) you would get the publishable results
XG(5.24.16): The order of experiments will be
(1) Exp. (C).
(2) If activation is spotted then Exp.(D).
(3) Exp. (O-1)(O-2)
(4) Exp. (E)
(5) Exp. (A-1) (A-2)
(6) Exp. (B-1)(B-2)
RC (5/23): It appears you have largely omitted FdL studies from the plan with DHP.So what happened to all the FdL studies you mentioned below and the standards you bought?There is no commentary on how you might provide some insight into why FdL gives false positives with DHP based eg on your control studies. Do you now plan to provide this commentary only for sirtainty?
You are planning to use only sirtainty, even though sirtainty was never used with SIRT3 in the publications?Sirtainty might be better than FdL for non-autofluorescent compounds, but what is the point of using it here?Is your hope that sirtainty will not provide the false positive with SIRT3? Or, are you proposing to use sirtainty because it uses unlabeled peptide? Because it is more reproducible?Do you intend to combine your old FdL results with the new sirtainty results for publication? If so, are you satisfied with your old control studies for publication of FdL results?
The point is to refute the published results for DHP/SIRT3. If you use other substrates (e.g. to show generality of results) or assays, it would need to be clear why and would ideally be in addition to the ones published.You mentioned that the reaction may not be driven by H3K9 for SIRT3. Did you choose it rather than p53 because it was used in the JMC2015 paper (verify), though not with SIRT3?In initial rate studies, a higher concentration of DHP than EC1.5 may be used for the reasons noted below. You did not account for or mention that under Alok's old results.
It also appears there are no initial rate studies with fluorogenic assay. This might be ok, but commentary must be provided on why and the HPLC conditionswould need to directly match those with fluorogenic in at least the endpoint assays. You did not highlight the directly conditions, although you previously planned torun HPLC and FdL on the same samples, as I recall.
Finally, there is no indication of the time required. I would need to know when each publishable result will be obtained.
All the above points should be addressed carefully with an eye toward publication.
I may not have time to provide further input at this point, esp since there is little commentary on the reasoning within the context of the stated goals. Please think carefully before doing experiments that take a month and result in nothing publishable. This cannot be sustained.
RC (5/22): --Following are some experimental priorities of our current work: -All published results must use in house enzyme.
XG(5.23.16): Enzo SIRT3 will be used for SIRTainty assay at the first set since it gives better activity.RC: For publication or not? See my comments above. Do you mean to publish both? That may be ok, but be aware of the time required.
1.
refuting honokiol activation for some substrates(not necc all; further mech studies as warranted) -With respect to your work with FdL, since Honokiol was not reported with FdL we would not need to compareto FdL results to refute. - Alok will be doing initial rate studies with p53or mnsod peptide first without honokiol then with-Since your FdL results with honokiol werenot always consistent with Hplc, we won't use FdL for publication of initial rate assays unless we see consistent effects in future with new substrates. This is ok sincewe primarily need to do two sets of initial rate exptsfor honokiol - with, without honokiol - and this can be handled on Hplc.
XG (5.23.16)
Honokiol/in-house SIRT3 activation experiment (2-3 days)
[In-house SIRT3]=10 ul
[AceCS2 peptide or H3K9]= ?
[NAD+] = ?
[Honokiol] = 0, 10, 50 uM
% DMSO = 1%
37oC
60 min
Note: [NAD+] and [AceCS2 peptide or H3K9] need to be determined based on Km, Vmax results for in-house enzyme. Also, need to know which peptide substrate will be more efficient for in-house SIRT3 on deacetylation reaction. In other word, which peptide substrate produce bigger product peak.
RC: I didn't necessarily mean that we need to do substrates beyond MnSOD with honokiol at this time. The comment above was a general comment about our work including Alok's work on MnSOD. I cannot post repeatedly on both pages.Note that any publication must show initial rate results.So, how are you planning to publish this result if it is negative?Is the goal here to just provide results with another substrate to demonstrate generality and complement Alok's endpoint results? In any case, why AceCS2 or H3K9 and not p53?Should these other honokiol substrates be tested here or later in the schedule? Provide your comments.
2.
refuting dhp activation -However with dhp we should compare Hplc to FdLunder the same conditions to refute. This is related to yourproposed expts. -Any initial rate expts with FdL substrate will onlybe usable on context of dhp refutation. For dhpone may compare the FdL substrate with FdL and Hplcassays.-You should assume that your sirtainty expt with dhp will be consistent with publication but inconsistent with Hplc.Will need to be repeated w Hplc for refutation. -For purpose of publication, the priority here would beto compare dhp initial rate studies with Hplc and FdLor sirtainty and based on your more rigorous analysis of controls/individual components/ standard curves provide some intuition as to why the Fluorogenic assays provided false positive results. However you should notaim to solve this problem, since the main goalis refutation along with some explanation that increases reader confidence that we are right. -In addition to refutation we may get some info onthe mode of inhibition of sirt by dhp. -You should use at least the Ec1.5 used in the papers. If you want to use a concentration somewhat higher thanEc1.5 for dhp that were also reported to show activation you can, in order to make sure we see some effect on initial rate parameters.
XG(5.23.16)
Exp. (A)
SIRTainty assay to evaluate DHP1c/SIRT1 activation (2015 JMC)
[SIRT1]=5U
[H3K9 peptide]=25 uM
[NAD+]= 200uM
[DHP1c]=50uM
30min
Ex/Em= 420nm/460nm
RC: I am going to assume 200uM NAD was used in publication.
Exp. (B)
If the activation is spotted, repeat experiment will be performed on HPLC under same condition.
Stored the samples from Exp. (A) in -80oC and check if it’s possible to run in HPLC directly.
Exp. (C)
(C-1) SIRTainty assay to evaluate DHP1c/SIRT3 activation---both enzo and in-house SIRT3 should be tested.
[Enzo SIRT3]=5U
[H3K9 peptide]=250 uM
[NAD+]= 500uM
[DHP1c]=50uM
60min
Ex/Em= 420nm/460nm
(C-2) [In-house SIRT3]=10ul
[H3K9 peptide]=250 uM
[NAD+]= 500uM
[DHP1c]=50uM
60min
Ex/Em= 420nm/460nm
Here: Study of km and Vmax for In-house SIRT3 should be performed to decide the NAD+ and peptide concentration (SIRTainty? HPLC?).
Reason for this experiment: to check if the deacetylation reaction can be driven by the combination of in-house SIRT3 and H3K9 peptide.
Exp. (D)
Repeat Exp. (C-2) using HPLC. This also will extend to “
compare dhp initial rate studies with Hplc and FdLor sirtainty”
Reason for this experiment: to check using old HPLC the detection limitation. In other words, how big the product peak will be and determine what concentrations need to be used for initial rate experiments.
Exp. (E)
Initial rate experiments DHP1c/in-house SIRT3 using HPLC
(E-1) Km, Vmax vs. NAD+
[In-house SIRT3]=10ul
[H3K9 peptide or AceCS2]=250 uM
[NAD+]= 0, 100, 250, 500, 1000uM
[DHP1c]=0, 50uM
%DMSO=0, 5% DMSO
0, 5, 10, 20, 30, 60, 120min
Note: 5min and 10 min time points need to be modified based on the peak area of product formation.
(E-2) Km, Vmax vs. substrate peptide
[In-house SIRT3]=10ul
[H3K9 peptide or AceCS2]=0, 25, 50, 100, 250, 500 uM
[NAD+]= 1500uM
[DHP1c]=0, 50uM
% DMSO= 0, 5%
0, 5, 10, 20, 30, 60, 120min
RC: Establishing the viability of initial rate experiments with HPLC (including which conditions/concentrations are quantifiable) is imperative. See my comments on Alok's page. You are both responsible for providing an answer to this asap sinceit would drive the publications.
-Check aloks old results to see if he saw any inhibition
by dhp at those concentrations on Hplc .The only concern is that dhp may only weakly bind to sirt at those concentrations and there may be little effect
Alok’s previous results on DHP1c/Enzo SIRT3
[NAD+]=500uM
[FdL2 peptide] =250uM
[SIRT3]=5U
5% DMSO
37oC
60min
% product formed in 0, 10, 25, 50 uM DHP1c are 2.12%, 2.06%, 1.84%, and 2.11%.
3.
establishing a higher throughput assay with reliableresults compared to Hplc, in absence of autofluorescence
4.
for DMSO, the only remaining relevant study wouldbe MM initial rate study on Hplc to see its effect on MMparameters. Regarding your FdL/Hplc comparison you should assume you will get consistent results in absenceof DMSO and autofluorescence but the real challenge willbe in presence of DMSO since many drug screening effortswill need to use it. Demonstrating this will be usefulfor scaling up future work but not an immediate priority forpublication.-As part of the initial rate studies you should also run DMSO without dhp to show the effect of DMSO oninitial rate parameters.
XG(5.23.16): Please see Exp (E) listed above.
For the purposes of our papers, 1,2,4 are higher priorities in that order.
5.
What is the plan in future due initial rate characterization of new enzyme batches - which assay and substrate?
XG(5.23.16): For new batch of enzyme purified in house (scale up using FPLC), at least for the first three batches we need to test km, Vmax, Kcat. If the numbers are close enough, then we can random test for the further batches. If the numbers are not consistent, then (1) purification method modification is needed (2) keep test the newly purified batches till it’s reproducible.
The test method will be HPLC or reliable assay (SIRTainty or some assay we established in lab). The choice of the substrate will be one of the three peptides (p53, H3, and AceCS2) currently available in the lab. We need to find out which one has the best efficiency for deacetylation reaction with in-house SIRT3. When the proposed experiments have completed, we will get good idea how the assay set up.
-If you encounter any issues with reproducibility of FdL or sirtainty assays that interfere with the above plan you should immediately move to help alok finish his Hplc work with honokiol more quickly.
XG(5.23.16): OK
-Please edit your schedule accordingly showing when (approx what dates) you would get the publishable results
RC (5/22):
--Following are some experimental priorities
of our current work:
- refuting honokiol activation for some substrates
(not necc all; further mech studies as warranted)
- refuting dhp activation
- establishing a higher throughput assay with reliable
results compared to Hplc, in absence of autofluorescence
- for DMSO, the only remaining relevant study would
be MM initial rate study on Hplc to see its effect on MM
parameters
For the purposes of our papers, 1,2,4 are higher priorities
in that order.
-With respect to your work with FdL, since honokiol
was not reported with FdL we would not need to compare
to FdL results to refute.
-However with dhp we should compare Hplc to FdL
under the same conditions to refute. This is related to your
proposed expts.
-All published results must use in house enzyme.
-Any initial rate expts with FdL substrate will only
be usable on context of dhp refutation. For dhp
one may compare the FdL substrate with FdL and Hplc
assays. Alok will be doing initial rate studies with p53
or mnsod peptide first without honokiol then with
-What is the plan in future due initial rate characterization
of new enzyme batches - which assay and substrate?
-Since your FdL results with honokiol were
not always consistent with Hplc, we won't use
FdL for publication of initial rate assays unless
we see consistent effects in future with new substrates. This is ok since
we primarily need to do two sets of initial rate expts
for honokiol - with, without honokiol - and this can be
handled on Hplc.
-You should assume that your sirtainty
expt with dhp will be consistent
with publication but inconsistent with Hplc.
Will need to be repeated w Hplc for refutation.
-Regarding your FdL/Hplc comparison you should
assume you will get consistent results in absence
of DMSO and autofluorescence but the real challenge will
be in presence of DMSO since many drug screening efforts
will need to use it. Demonstrating this will be useful
for scaling up future work but not an immediate priority for
publication.
-For purpose of publication, the priority here would be
to compare dhp initial rate studies with Hplc and FdL
or sirtainty and based on your more rigorous analysis
of controls/individual components/ standard curves provide
some intuition as to why the Fluorogenic assays provided
false positive results. However you should not
aim to solve this problem, since the main goal
is refutation along with some explanation that increases
reader confidence that we are right.
-In addition to refutation we may get some info on
the mode of inhibition of sirt by dhp.
-You should use at least the Ec1.5 used in the papers.
-Check aloks old results to see if he saw any inhibition
by dhp at those concentrations on Hplc . The only
concern is that dhp may only weakly bind to
sirt at those concentrations and there may be little effect.
If you want to use a concentration somewhat higher than
Ec1.5 for dhp that were also reported to show activation
you can, in order to make sure we see some effect on
initial rate parameters.
-As part of the initial rate studies you should also
run DMSO without dhp to show the effect of DMSO on
initial rate parameters.
-If you encounter any issues with reproducibility of
FdL or sirtainty assays that interfere with the above plan
you should immediately move to help alok
finish his Hplc work with honokiol more quickly.
-Please edit your schedule accordingly showing when
(approx what dates) you would get the publishable results
RC (5/20):** Based on the recent email correspondence outcome I would like you to post a schedule here.
Note the very important point that you will need to contribute to some of the experiments relevant to papers in the near term,and as such since fluorogenic assays may not be usable in the very short term (until a suitable assay is developed, not clear how rapidly that will be possible - you can indicate), you may needto allocate some more time to HPLC. See the relevant tasks and discuss accordingly to establish your role.XG((5.20.16): Experiment I am currently working on / planning to do
- Validate DHP1c/SIRT1 activation experiment (2015 JMC) using SIRTainty assay. SIRTainty kit order has been sent out on 5.18.2016. Expecting to receive it early next week.
- Develop protocol for run FdL assay reaction sample on HPLC for direct comparison of results from both assay system
(1) Run individual component on Beckman HPLC (old)
i. NAD+
ii. NAM
iii. Peptide substrate (first FdL2 peptide)
iv. FdL standard
v. 1xDeveloper
vi. AMC
vii. FdL standard + 1xDeveloper
(2) If the a-f peaks are not overlapping, then run same sample from FdL assay directly.
(3) Choose the suitable component and run standard curve
3. Km, Vmax measurement for in-house enzyme (AE method). Plan to do two different batches to testy the consistency of the in-house SIRT3 performance.
- Firstly will use FdL
- Then modify HPLC conditions (time points, enzyme concentration, NAD+ and peptide substrate concentrations). This will benefit the initial rate study.
RC(5.12.16): Many answers to questions are still pending. I will not list them all here.To just list a few: summary of which FdL/honokiol experiments were repeated and what was found with HPLC analysis of honokiol activation vis-a-vis the FdL results,XG(5.12.16): we received bulk SIRT3 enzyme from Enzo yesterday. SM and XG aliquot them this morning. The inventory of the aliquots was created for tracking. XG start the FdL/Honokiol repeat experiments today using bulk enzyme.
RC: why HPLC was not used to evaluate DMSO effect,XG(5.12.2016): XG need to look through AU's report to find out the new HPLC test limitation to design the DMSO effect experiment. It will be followed right after Honokiol repeat experiment.
RC: This expt will be important -- see below.XG(5.16.16): XG had run the reactions on 5.13.2016. The samples were frozen into -80oC right after the completion of the reaction. The samples are running on new HPLC now. Will provide a report tomorrow.
XG(5.17.16): Validation of DMSO effect using HPLC result is attached.
Validation of DMSO effect by using HPLC.pptx
RC: how FdL can be used if there is autofluorescence from DMSO,XG(5.12.2016): we need to have assay buffer as background, % DMSO as positive control
•Background control: NAD+, peptide substrate, DMSO•Positive control: NAD+, peptide substrate, DMSO, enzyme•Compound control: NAD+, peptide substrate, the compound at the same concentration as its experimental counterpart. •Delta AFU_compound= Experimental well signals at all tested compound doses – compound control•Delta AFU_control = Positive control – background control•The % activation was derived from % Activity = DAFU_compound /∆AFU_control
RC (5/13): This is not enough for reasons we have discussed several times. Without proper standard curves, the extent of contribution from autofluorescence can change with FdL product concentration. Do you admit DMSO induces autofluorescence at 5%? If so, you should not waste time and money doing experiments that are inconclusive -- you need to convince us that your procedure is reliable.You need to validate the results/protocol with hplc asap (using new standards in your FdL protocol) if required.Or, if you can't, propose a new protocol/assay that can be used with DMSO asap. An assay will not be accepted unless it can verify hplc results. If you can't achieve this during this phase of the work, spend your time with (old) HPLC instead and achieve this afterwards.RC: At what concentrations of DMSO can you confirm that FdL works reliably (no false positives)? Have you cross-compared with HPLC to verify?XG(5.17.16): %DMSO in range 0 - 20% has been tested using both FdL assay and HPLC method. HPLC results shown that pmoles of product formed decreases as increasing %DMSO. under same conditions the FdL assay was carried out. Similar phenomenon was spotted.
RC: If not yet, which of your previous FdL/honokiol results have your cross-compared with HPLC results from Alok to verify?
XG(5.16.16): Experimental conditions for HPLC and FdL a little difference due to the detection limitation of HPLC. For example, under 10uM FdL2 peptide and 100uM NAD with10 uM honokiol experiment, XG used 5U SIRT3. However, 8U was used in HPLC experiment since the product peak was too small to quantitate. XG and AU discussed about the issue, two possible solutions (1) XG redo the experiment using 8U SIRT3; (2) AU rerun reactions using 5U SIRT3 for longer incubation time. Because FdL assay is faster, solution (1) will be proceeded.
RC: what enzyme is being used for what,XG(5.12.16): Bulk SIRT3 enzyme (lot#: 05041621) will be used for FdL validation/ initial rate and HPLC work. SM in-house enzyme will be used for repeat Honokiol/SIRT3 initial rate experiments.
comparison of new hplc quantification of unlabeled peptides with old hplc data and how things may change with in-house enzyme,whether initial rate expts will be possible with hplc, what/when new hplc experiments will be done with honokiolRC(5/13): Not at all clear what this means. "used for FdL validation/ initial rate and HPLC work" basically means everything? So this is not informative. Initial rate studies should be done with in-house enzyme.More importantly, no one has properly addressed my postings on Alok's page indicating that the schedule for all experiments needs to be aligned so the order and dependency is the way I originally intended.And, I had mentioned that the posting on this page was not a comprehensive list of the unanswered questions.Given the input I have provided here, you and Alok will need to arrange schedule accordingly to get the most important results i have asked for in the right order.
I don’t want to see weeks of FdL work later rejected by HPLC or weeks of HPLC initial rate experiments in presence of modulator done under conditions that were not ideal due to insufficient endpoint experiments done in advance
XG(5.16.16): Good point. Currently XG is doing validation (1) DMSO effect using HPLC; (2) under AU's condition (8U SIRT3) for honokiol activation. As soon as (1) and (2) finished, we will know (A) if %DMSO provide false positive results in FdL assay. (B) if Honokiol activates SIRT3 using FdL2 peptide. If YES for DMSO, then new assay system is needed. SIRTainty assay was used for DHPs study. In their 2015 JMC paper, DHP1c was tested using SIRTainty assay and activation of SHP1c on SIRT1 was detected. We may want to order SIRTainty kit to repeat (a) DHP1c on SIRT1 (b) if any DMSO effect in this assay system. If No for (B), MnSOD (peptide/ protein) need to be ordered
RC: bearing in mind that initial rate experiments with FdL/honokiol may depend on HPLC validation,XG(5.12.16): Yes.
RC: whether there is any time left in sudipto's schedule for any hplc expts, etc. The current work being done in the lab and why is largely unclear.
SM(5.12.16): Today I am finishing up the last batches of Sirt3 purification for XG's and AU's experiments. I will be continuing the literature search for Sirt1 and Thermophoresis tomorrow.Monday (5/16) I will measure the activity of all the newly purified Sirt3 batches and begin conditioning the new column for HPLC.RC: You mentioned you would post a schedule for truncated SIRT3 purification.SM(5.16.16): I have obtained a quote from GenScript (https://www.genscript.com) for the truncated Sirt3 (118-399) construct:GenSript_Quote_Sirt3(118-399)+pET14b.pdf
- The total cost for the construct will be $294.39
- The estimated delivery time is 8 days
- Once the order arrives, a plasmid prep will be done to obtain sufficient amount of plasmid and the sample will then be sent for sequencing (total time ~ 1 week).
- If the sequencing results come back as correct, then the plasmid will be transformed into Rosetta 2 (DE3) cells for overexpression (2 days).
- Optimizing overexpression conditions - IPTG concentration, temperature, growth time, etc (~ 1 week).
- Large scale overexpression (~ 1 week).
- Purification - need to find a workable protocol and then optimize it (~2 weeks).
Should I go ahead and order the truncated Sirt3 construct?
RC(5.9.16): See today's posting on Alok's page.Comments and concerns regarding the experiments you listed below are mentioned there.Also comments regarding other tasks and questions are listed.RC (5.6.16):Today, I would like to see a list of expts that will be done by all 3 lab members next week,as well as a lists of all pending questions listed that will be answered next week.XG: Monday XG and SM will test the activity of SIRT3 new samples from Enzo bulk order and prepare report. If the result pass the test, we will request the shipment for bulk order. The SIRT3 will be ideally shipped to lab Tuesday. Then the scheduled experiments will proceed. (1) %DMSO activation/inhibition on SIRT3(2-3 days)
[SIRT3] = 5 U; [NAD+] = 100, 1500uM; [FdL2 peptide] =25, 100, 250 uM; % DMSO = 0, 1%, 5%, 10%, 20%, 30%, 35%, 40%, 50%; Time points = 0, 60 min; Temp. = 37oC
RC: The issue here is not just to repeat the DMSO experiments but to answer the question as to whether DMSO contributes autofluorescence that can lead to false positives
I asked you make the assay systematic and reliable without false positives or choose one that is (see below). See also the HPLC validation task for this.
XG: we need to repeat the experiments to check if the previous results is consistent. Then the contribution of DMSO
(2) SIRT3 activation by Honokiol using Enzo enzyme / Sudipto in house enzyme (2-3 days)
[SIRT3]=5U/reaction;[NAD+]=100, 1500 uM;[FdL2 peptide]=10, 25, 50, 100, 200, 250 uM;[Honokiol]=0, 10uM;% DMSO= 1%;Time points= 0, 60 min; Temp.= 37oC
If by the end of next week the Experiment (2) still not finish, then will push to week after.
XG( 5.2.16): Thank you for the comments. Sutipto and I are working on the measurement of the Bulk SIRT3 enzyme sample activity this morning. We need to send data to Enzo so that the bulk SIRT3 can be received early this week for experiments. I will response the questions right after the activity measurement.
-- DMSO-Consistency or lack thereof with old results: You previously indicated that DMSO activates SIRT3. Here we see inhibition at 1% (though lower [peptide]).-Dependence on peptide concentration: it appears 1% DMSO inhibitory effect diminishes at higher [peptide]. This may be due to DMSO causing increase in peptide Kd, which does not have much effect near saturating peptide concentrations. Does DMSO activate at saturating peptide?-Increase in AFU at 0 min (5% DMSO).
XG: Previous experiments
[SIRT3]= 5U
[NAD+] =1500uM
[FdL2 peptide]= 100uM
% DMSO = 0 – 50%
Time points= 0, 60 min
The SIRT3 activation by the addition of DMSO was detected at 10%, 20%, 25%, and 30% DMSO. The signal goes down most likely due to the denaturation of SIRT3 by high% DMSO.
To confirm, the above experiments need to be repeated twice. 1% and 5% DMSO should be included in the experiments. Experimental condition is listed
[SIRT3] = 5 U
[NAD+] = 100, 1500uM
[FdL2 peptide] =25, 100, 250 uM
% DMSO = 0, 1%, 5%, 10%, 20%, 30%, 35%, 40%, 50%
Time points = 0, 60 min
Note that some of the conditions are overlapping with Honokiol experiments. Will combine them to save time.
Consistency with previous claim that there is no relevant autofluorescence from DMSO. Never highlighted before. Concerning.
XG: DMSO emits light at wavelength ~354 nm.
RC: This however is not an answer to the question of why you previously indicated DMSO does not contribute to baseline fluorescence at the measured wavelength for FdL
and that it does not introduce autofluorescence issues.
Need an answer to determine whether we can keep using the FdL assay with current controls for honokiol/DMSO in future work.
-- Standard errors of reported quantities/reproducibility of honokiol results:-Data on % activation shows significant variations both between peptide concentrations and between repeats of the same experiment (e.g. 200uM peptide).-You said you did triplicate experiments on activators like resveratrol. Present the cvs and standard errors of those measurements (raw data) as well as the % activation in that case.
XG: Yes. Triplicate experiments were done for resveratrol activation experiment. The % activation, stdv, and cv% were presented on slide 3/SIRT1 activation by Resveratrol_04.18.16.ppt.
Report on whether the variations observed in the honokiol experiments lie within those standard errors (raw data and % activation).
XG: The duplicate experiments were only done on the reactions where activation was detected. The rest of the experiments were done once. --Various typos in the ppt (100 vs 1000uM, FdL1 vs FdL2, etc)
XG: Typos have been corrected.Honokiol-SIRT3_04.29.2016.pptx--Next expts: no point in doing DHP now with FdL since you have not yet resolved issues with controls esp for cases with autofluorescence. After confirming above that the FdL assay can be used with current controls despite DMSO effects. Instead repeat the honokiol experiments again (100uM NAD, 1500uM NAD). Determine whether you can reproduce the same trends, esp that of 100uM NAD+ showing some apparent activationand 1500uM NAD+ showing only apparent inhibition-If you can validate the results and Alok also sees consistent results with HPLC, you should then do a quick repeat with Sudipto's enzyme (if it can be done within a day) and then focus on Km,vmax determination in presence of honokiol (10uM). This should be done under two conditions: a) saturating NAD+, non-saturating peptide; b) saturating peptide, nonsaturating NAD+. MM fittings for these should be provided. Consider presence/absence of DMSO as well. Propose the schedule for these experiments.
XG: Proposed schedule
(1) %DMSO activation/inhibition on SIRT3 (2-3 days)
[SIRT3] = 5 U
[NAD+] = 100, 1500uM
[FdL2 peptide] =25, 100, 250 uM
% DMSO = 0, 1%, 5%, 10%, 20%, 30%, 35%, 40%, 50%
Time points = 0, 60 min
Temp. = 37oC
RC: The issue here is not just to repeat the DMSO experiments but to answer the question as to whether DMSO contributes autofluorescence that can lead to false positives
I asked you make the assay systematic and reliable without false positives or choose one that is (see below). See also the HPLC validation task for this.
(2) SIRT3 activation by Honokiol using Enzo enzyme / Sudipto in house enzyme (2-3 days)
[SIRT3]=5U/reaction
[NAD+]=100, 1500 uM
[FdL2 peptide]=10, 25, 50, 100, 200, 250 uM
[Honokiol]=0, 10uM
% DMSO= 1%
Time points= 0, 60 min
Temp.= 37oC
(3) KmNAD+, VmaxNAD+ measurement in the presence of 10uM honokiol
a, Saturating NAD+, non-saturating FdL2 peptide (2 days)
[SIRT3]=5U/reaction
[NAD+]=1000, 2000, 3000, 4000 uM
[FdL2 peptide]=10 uM
[Honokiol]=0, 10uM
% DMSO= 1%
Time points= 0, 10, 20, 30, 60, 120 min
Temp.= 37oC
b. Non-saturating NAD+, saturating FdL2 peptide(2 days)
[SIRT3]=5U/reaction
[NAD+]=125, 250, 500, 750 uM
[FdL2 peptide]=250 uM
[Honokiol]=0, 10uM
% DMSO= 1%
Time points= 0, 10, 20, 30, 60, 120 min
Temp.= 37oC
c. Higher Honokiol concentration(2 days)
[SIRT3]=5U/reaction
[NAD+]=?
[FdL2 peptide]=?
[Honokiol]=0, 50 or 100uM
% DMSO= 1%
Time points= 0, 10, 20, 30, 60, 120 min
Temp.= 37oC
Report on the effects of honokiol on Km,vmax for a,b) above. Thereafter, you may need to do one more higher [honokiol] to check for partial or complete inhibition, along with a mixed inhibition fitting. If Alok's results are not consistent, indicate that to me and wait for my response. Note these experiments will constitute standard mixed-inhibition type studies to identify which kinetic parameters are being modulated by honokiol, prior to doing any further mechanistic analysis.(Related, please point me to where you originally reported Km,vmax for SIRT3 with and without DHP [MM fittings] prior to embarking on our initial rate studies including both DHP/NAM.)
--Systematization of non-HPLC based assays.-Update on status of ordering of the synthesized deacetylated peptideXG(5.6.16). The quote request for the following four peptide had been sent out.
The quotation is back and the delivery time is 3-4 weeks.
quote_105296-peptide10mg.pdfquote_105297-peptide25mg.pdf
RC(5.6.16): What you mean the four peptides? Have we ever referred to 4 FdL peptides before? Post your answer on wiki.
XG(5.6.16) :
The four peptides include
-- the FdL2 peptide named as P53317-320-Ac-AMC (no.4 in the table), which is acetylated peptide with AMC dye conjugation. By calculation, 25mg of N.o4 peptide, will make 8.5 ml of 5mM FdL2 peptide solution. It costs $275.2 for synthesize. We currently purchase FdL2 peptide from Enzo as $178 per 100ul of 5 mM solution.
-- For the purpose of suitable standard, it should be deacetylated FdL2 peptide, which named as p53317-320-AMC (deacetylated peptide with AM dye conjugation, no.3 in the table). When 1x developer II applied, the fluorescence released and AFU will be readout.
-- No. 1 and 2 are deacetylated and acetylated FdL2 peptide without AMC. The reason No. 1 and 2 were included are, (1) use No1 for the reaction to validate if AMC has effect on honokiol/SIRT3 results; (2) No. 2 deacetylated peptide can be used for standard curve in HPLC.
Now we have an rough idea from the quote. We can decide not to purchase all of them.
Please advise.
XG(5.9.2016): Please advise if we should order.
RC: Yes, we can proceed to order the set.Of course, the AMC conjugated peptides are most important to work with to start. So far, we have not confirmed that honokiol activates SIRT3 with AMC FdL peptide.Hence there is no immediate need to validate the AMC effect on honokiol activation with this peptide, though we would do so later should activation be confirmed. The presence/absence of tag may have an effect regardless.Presenting the experimental plan for proper FdL controls with the tagged peptides is the priority. We need to establish whether having the proper standards can avoid false positives in FdL assay.This is esp imp if solvent like DMSO can lead to false positives/autofluorescence.Explain the reasoning behind your proposed experiments with reference to our previous discussions.We are doing p53 experiments and possible MnSOD on HPLC as well for an untagged peptide.BTW, is there any reason to believe that the FdL assay can work with only their peptides above?XG(5.9.16): Initial rates of deacetylation were determined for a series of fluorogenic acetylated peptide substrates (Enzo SIRT3 assay manual). Among them, FdL2 peptide was the substrate deacetylated .
Concurrently, I am still waiting for more systematic sirtuin assay analysis (inclluding Sirtainty and others) as noted below, vis-a-vis development of proper FdL controls. We want to know which assay can provide the most rigorous controls without false positives. E.g., I asked questions about whether there would be similar issues with standards/controls with Sirtainty, etc.
XG(5.9.16): It needs a lot literature search. I am still working on this.
For it to serve as a reliable higher throughput method, you would need to establish consistency between the proposed protocol and HPLC in almost all cases - e.g., no false positives, similar kinetic parameter estimates, etc.-Update on the questions regarding alternate assays like Sirtainty and their background/standard curve issuesXG(5.11.16): The comparison of SRITainty and Enzo assay was listed in ppt.
SIRTanity.pptx
Still working on other alternated assays.
-Conclusions regarding appropriate controls to use with FdL, or with other assays identified as being more suitable.
Provide schedule for all the above. Since they are faster than HPLC, they should be done 1-2 weeks faster, so as to leave tim for further mechanistic studies with honokiol with the remaining timeshould they be warranted.
RC:You may personally work on one of the HPLCs to accomplish some of the validation tasks if you have available time and the others have not been assigned or do not have time to finish those tasks. You are not tied only to FdL.
XG: Sure. Since FdL is faster than HPLC, I can jump to HPLC when I have available time. When I reach that point, I will discuss with Alok about what validation tasks need to be done.
RC: Please comment on how other authors, such as those of the Ex-527 SIRT3 PNAS paper, and Sir2Tm authors who estimated kcat, were able to estimate kcat despite activity/denaturation issues (not just vmax).
XG: In Ex527-SIRT3 PNAS 2013 paper (supporting information Figure S1A), kcat, Vmax were reported as a Table. It was claimed in Material and method section, 2.5uM for Sir2Tm and Sirt3 were used for kinetic study. They use full length Sir2Tm and his-tag Sirt3 (114-380). The specific activity (nmol/min/mg) was used as Vmax. The calculation see below)
shown [E]Total was close to 2.5uM as mentioned in the paper. This indicated that the authors assume the total protein are active.
RC: You should both follow up on questions regarding comparison of specific activity from these papers to ours.
Discuss with Sudipto the purification protocol used in PNAS in this regard and ask him to compare his.
XG: Note that the sequence of SIRT3 used for kinetic study is 114-380, which is different from ours (102-399). Sudipto will compare the purification protocol soon.
SM: The main difference between the PNAS protocol and AE protocol is the length of the Sirt3 gene used for expression. In our case, we use the 102-399 version, whereas they have used the 114-380 version. The other difference is that they used BL21(DE3) Rosetta 2 cells for expression whereas we use Arctic Express. Their Sirt3 is soluble in the Rosetta 2 cell line because from literature it seems that the shorter versions (118-399 or 114-380) are more soluble than the 102-399 version. Other than that, the purification procedure is similar (they also use His-tag based purification).
RC: We have discussed solubility of different constructs in the past. Is solubility explicitly mentioned?
I think the paper Guan id'd with high purity/spec activity also used our construct in one case?
Do you think we could get significantly higher specific activity and purity (hence improving assumption that all enzyme is active) with the shorter version?
You should both elaborate on this since it is important.
How long would it take to try expression and purification of the shorter version? Relevant orders could be made (Guan - follow up when time permits if this looks promising) and used later.
What about for the other sirtuins - is this problem mentioned?
SM: The solubility issue is not explicitly mentioned in literature, but there are several indications that the 102-399 construct is less soluble than the truncated version. For example in the Jin (2009) JBC paper, the truncated version (118-399) was expressed in BL21 (DE3) cells (used for soluble proteins), while for the 102-399 construct, they had to coexpress the pG-KJE8 chaperone protein along with the Sirt3 construct in BL21 (DE3) cells. Since the chaperone proteins are used for improving solubility, it could be inferred that the 102-399 construct was less soluble than the truncated version. They also mentioned that they could not crystallize the 102-399 protein.
Upon comparison of the activities of the two constructs, they found that it was similar. However, they mention that their result is in disagreement with Steegborn (2008) JMB result where they found that the specific activity of truncated Sirt3 (114-399) was ~50-fold higher than 102-399.
RC: Ok then there is a tradeoff between accurate representation of the activity of the full length protein and the solubility.We should probably proceed in parallel with truncated version expression and purification esp given that was used for crystallization (hence for modeling).Please present a timetable for this (see other questions above).SM: For the truncated version, PCR, cloning, sequencing and selection of the correct clones/constructs would take about 4 weeks, if all goes well. Once we have the right construct, expression and small scale purification (without optimization) would take another 2 weeks.RC: Place the necessary orders.See my other question above.BTW, when you say specific activity of truncated SIRT3 is 50 fold higher, couldn't that just be because of the denaturation of the protein during purification of the longer construct? Isn't that expected based on what we saw in our specific activity measurements?And is this likely the reason they were able to provide kcat?Any evidence that 114-380 is more soluble than 114-399?Is Enzo using 102-399 SIRT3?
SM: Yes it is possible that the specific activity of the truncated Sirt3 is higher because of denaturation of the longer construct during purification. However, this difference in activity of the two constructs has only been reported in one paper (Steegborn, 2008, JMB). We are not sure if that is likely the reason they were able to provide kcat.
There is no reported evidence from literature that the 114-380 is more soluble than 114-399. Yes, Enzo is also using 102-399.
XG(5.9.16) Yes. Enzo SIRT3 is 102-399. I have calculated % product formation for Enzo and JBC enzymes. I will include in-house enzyme when I get time. Please see ppt.
Enzyme activity comparison_5.9.2016.pptx
Guan, this may be quite important please look into it and let me know once we are ready with the reagents so we can decide whether to proceed.Follow up on the other questions about specific activity comparison to lit in this regard.
RC: Please answer the key questions regarding whether we expect that truncated SIRT3 has higher specific activity because it did not have to be subjected to denaturating conditions or because it was not complexed to chaperonin.If we expect to be able to report kcat with the truncated protein, and if it was also used for the crystal structures we are using, it is important to proceed with purification of that protein as well. A schedule would need to be presented whereby we couldhave it within a month and a half or so. We would continue using AE enzyme in the meantime without reported kcat and then compare the two enzymes later.
The specific activity comparison to in-house is also critical.
SM: The truncated Sirt3 seems to be more soluble (and hence easier to purify), from literature. Since the more the number of purification steps (or denaturing conditions), the less the activity of the enzyme, it is possible that the difference in activity between the two constructs is because of their solubility in the cytoplasm of the cell.
I will work on a detailed schedule on this and update.
4.29.2016
Honokiol-SIRT3_04.29.2016.pptx
4.18.2016
SIRT1 activation by Resverstrol_04.18.16.pptx
DHP1c intrinsic fluoresence controls.pptx
4.4.2016_ Updated Schedule
(1) Saturating FdL1 peptide concentration for SIRT1----------------2 days
[NAD+]= 500, 3000uM
[FdL1 Peptide]= 0, 25, 50, 100, 200, 300, 400, 425, 450, 470, 500, 600, 700, 800, 1000uM
[SIRT1]=1U
Time points= 0, 60 min
37oC incubationFdL1_FdL2 peptide saturating concentrations.pptx
(2) DHP1c EC1.5 for SIRT1 under new condition-----------2 days
[NAD+]= 500 uM
[FdL1 Peptide]= 250 uM
[SIRT1]=1U
[DHP1c]=0, 1, 10, 50uM
% DMSO= 5%
Time points= 0, 60 min
37oC incubationDHP1c-SIRT1_04.07.2016.pptx
(3) Honokiol EC1.5 for SIRT3 -----------2-3 days
Based on Alok’s “Honokiol solubility” results, this set of experiments will be performed in the presence of 5% DMSO. The respective standard curves will be performed as well.
[NAD+]= 500 uM
[FdL1 Peptide]= 250 uM
[SIRT3]=5U
[Honokiol] = 0, 10, 100, 200uM
% DMSO=5%
Time points= 0, 60 min
37oC incubation
Honokiol-SIRT3_04.08.2016.pptx
RC (4/4/16): You need to consider whether the 5% DMSO or DHP was the primary source of intrinsic fluorescence issues in past experiments.If DMSO had a significant effect, this will cause issues with FdL assays of the planned honokiol study as well.Then other approaches to improvement of solubility that reduce these background effects may need to be considered for FdL based experiments.HPLC should not have this problem.XG (4.4.16): The primary source of intrinsic fluorescence was mainly from DHPs because at 450nm DHPs emit lights which overlap with those from fluorophore produced by deacetylated peptide. The emission wavelength for DMSO is 350-354nm. Of course, it will be ideal if we can reduce %DMSO used in honokiol solution. I will discuss with Alok to test honokiol solubility in lower % DMSO.
3.28.2016
DHP2c_03.28.2016.pptx
Time series of product formation vs. time_DHP1c.pptx
New in-house protein
Protein purification _Urea_03.18.2016.pptx
Updated Schedule (3.8.16)
03.03.16-03.09.16 Cell culture 8X200ml
03.09.16 – 03.11.16 purification
03.14.16 characterization of purified enzyme (concentration, activity)
03.15.2016 Km, Vmax measurement for new purified enzyme (Before continuing the initial rate exp. I need to confirm the newly purified enzyme is good. Plus we can get the km, Vmax values.)03.16.2016 – 03. 17.2016 Finish the initial rate experiments.
RC: I assume this refers to Km,Vmax on combined batch. What about 3.14 characterization?For completeness please specify the series of concentrations in same format as below.XG: 3.14.16 will be doing characterization of purified enzyme from different batches. The combination of protein from different batch will be done within the batches with similar concentration and activity.
RC: How many batches will be prepared? How many need to be combined?XG: 3 batches. Depends on the concentration and activity, will decide at that point.
RC: How does this connect with plan for Km,Vmax reproducibility analysis? I.e., if you are combining batches before checking Km,Vmax, how will you check reproducibility?
XG: Good point. To check the reproducibility from batch to batch, the Km and Vmax need to be measured individually before combining. One set experiment without duplicate needs 40 reactions plus standard curve which need one day per batch. The updated schedule is listed below
03.14.16 characterization of purified enzyme (concentration, activity)
03.15.2016-03.17.2016 Km, Vmax measurement for new purified enzyme from each batch (Before continuing the initial rate exp. I need to confirm the newly purified enzyme is good. Plus we can get the km, Vmax values.)
03.18.2016 and 03. 21.2016 Finish the initial rate experiments.
03.22.2016 Data analysis and model fitting If the data cannot match.
03.23.2016 and 03.24.2016 redo previous experiments
03.25.2016 Data analysis and model fitting
03.28.2016 – 03.29.2016 Higher NAM concentration modification using newly purified enzyme
03.30.2016 – 03. 31.2016 Initial rate experiments for higher NAM concentration [NAD+] = 375, 750, 1500, 3000 uM [NAM] = 0, higher concentration [DHP2c] = 0, 25uM Time points: 0, 5, 10, 20, 30, 60, 120 min
04.01.2016 Data analysis/model fitting
04.04.2016 – 04.07.2016 Duplication [NAD+] = 375, 750, 1500, 3000 uM [NAM] = 0, 50, 100, higher concentration [DHP2c] = 0, 25uM Time points: 0, 5, 10, 20, 30, 60, 120 min
04.08.2016 Data analysis / model fitting
03.18.2016 Data analysis and model fittingIf the data cannot match,03.21.2016 and 03.22.2016 redo previous experiments
03.23.2016 Data analysis and model fitting
03.24.2016 – 03.25.2016 Higher NAM concentration modification using newly purified enzyme
03.28.2016 – 03. 29.2016 Initial rate experiments for higher NAM concentration
[NAD+] = 375, 750, 1500, 3000 uM; [NAM] = 0, higher concentration;[DHP2c] = 0, 25uM; Time points: 0, 5, 10, 20, 30, 60, 120 minRC: By 3.18, please consult with me on choice of higher [NAM].XG: OK
03.30.2016 Data analysis/model fitting
03.31.2016 – 04.05.2016
Duplication [NAD+] = 375, 750, 1500, 3000 uM [NAM] = 0, 50, 100, higher concentration [DHP2c] = 0, 25uM Time points: 0, 5, 10, 20, 30, 60, 120 min04.06.2016 Data analysis / model fitting
RC: What is meaning of "and 03.14.2016".I believe the duplication experiments are identical to the complete set of initial rate experiments above (including higher [NAM].XG: It's Typo
Signal to noise comparison_3.4.16.pptx
Updated effects.ppt
updated DMSO effects _2.26.2016.pptx
DMSO effectsDMSO effects_2.4.2016.pptx
02.17 - 02.19.2015
- Harvest cells and store in -80oC for Sudipto use.
8 X 200ml of SIRT3 culture were harvested and stored in -80oC. 500ul of samples were taken from each bottle plus un-induced sample. Run SDS-page gel for protein expression level.
* Endpoint (2hours) control Exp. using Enzo enzyme. Calculate theoretical amount of product formation.
* SI preparation first draft. The underline parts are from Dr Raj’s previous comments and emails. References were edited using Endnote.
SI preparation for standard curve_2.19.2016 update.docx
Updated Schedule (2.15.2016)
02.15.2016
- Aliquot buffers for future experiments.
- Continue working on SI preparation
- Inoculate the culture from glycerol stock at 37oC overnight
- Transfer overnight culture to 200mL at 37oC then IPTG induction at OD~ 0.6, 30oC overnight
- Discuss with Alok about estimation of how many batches of enzyme is needed for HPLC
- Hopefully finalize the first draft of SI preparation and send to Dr Raj to review
SI preparation for standard curve_2.11.2016 update.docx
02.17 - 02.18.2015
- Harvest cells and store in -80oC for Sudipto use.
- Endpoint (2hours) EC1.5 measurement for DHP2c using Enzo enzyme. Calculate theoretical amount of product formation for 2 hours. Will check the if the standard curve need to be extended to higher [ ].
02.22.2016 – 02. 23.2016 Finish the initial rate experiments.
02.24.2016 Data analysis and model fitting If the data cannot match.
02.25.2016 and 02.26.2016 redo previous experiments
02.29.2016 Data analysis and model fitting
03.01.2016 – 03.02.2016 Higher NAM concentration modification using newly purified enzyme
03.03.2016 – 03. 04.2016 Initial rate experiments for higher NAM concentration [NAD+] = 375, 750, 1500, 3000 uM [NAM] = 0, higher concentration [DHP2c] = 0, 25uM Time points: 0, 5, 10, 20, 30, 60, 120 min
03.07.2016 Data analysis/model fitting
03.08.2016 – 03.11.2016 and 03.14.2016 Duplication [NAD+] = 375, 750, 1500, 3000 uM [NAM] = 0, 50, 100, higher concentration [DHP2c] = 0, 25uM Time points: 0, 5, 10, 20, 30, 60, 120 min
03.15.2016 Data analysis / model fitting
Updated Schedule (2/12/2016)
02.15.2016 Km, Vmax measurement for new purified enzyme (Before continuing the initial rate exp. I need to confirm the newly purified enzyme is good. Plus we can get the km, Vmax values.)
02.16.2016 – 02. 17.2016 Finish the experiments.
02.18.2016 Data analysis and model fitting
If the data cannot match.
02.19.2016 and 02.22.2016 redo previous experiments
02.23.2016 Data analysis and model fitting
Updated Schedule (2/8/2016)
RC (2.5.16): Note that we may add a higher [NAM] at the end; I will advise later.
RC (2.5.16): We may also later (after experiments are underway) decide whether to increase the number of duplicate experiments.
RC (2.5.16): Regarding mixed fitting, you should let me know before you do the final fittings since I will have some comments.
*Purified enzyme initial rate study
[NAD+]=375, 750, 1500, 3000 uM
[DHP2c]=0, 25uM
[NAM]=0, 50, 100 uM
% DMSO = 0%
Time points: 0, 5, 10, 20, 30, 60, 120 min
8*2*4*3= 192 reactions----------4 days experiment 1 day data analysis (Feb 08-12, 2016)
- SI draft should proceed concurrently.
Updated Schedule(2/1/2016) EC1.5 measurement with purified protein
[DHP2c] = 0, 10, 25, 50, 75, 100, 150, 200, 300, 400 uM
% DMSO = 0, 0.2%
Time points: 0, 60 min
9*3*2*2=108 reactions----------2 days (Jan 18- Jan19, 2016)
Standard curve for EC1.5 calculation
[DHP2c] = 0, 10, 25, 50, 100, 200, 300, 400 uM
% DMSO = 0, 0.2%
9*8*3=216 reactions -------- 3 days (Jan 20-Jan 22, 2016)
@One more set of standard curve for inter-day variation evaluation.
@Try higher [Standard] and test linearity
If the 0.05% can do then omit 0.5% one.
Standard curve at different DHP2c w_o DMSO_1.22.2016.pptx
Standard curve
[DHP2c] = 0, 50, 100, 200, 400, 500uM, 1mM, 2mM
% DMSO =1%
Endpoint enzymatic reaction to test % activation at following condition
[DHP2c] =0, 50, 100, 200, 400, 500uM, 1mM, 2mM
% DMSO =1%
Time points: 0, 60min
8*8*2+2*8*4= 192 reactions ------3 days (Jan 26 – Jan 28, 2016)
Standard curve at different DHP2c w_o DMSO_1.27.2016.pptx
New Enzo enzyme initial rate study
[NAD+]=375, 750, 1500, 3000 uM
[DHP1c]= 0, 50uM
% DMSO =0, 5%
Time points: 0, 5, 10, 20, 30, 60, 120 min
8*2*4 +4*8 = 96 reactions---2 days experiments and 1 day data analysis (Jan 29, Feb 1 -2, 2016)
Purified enzyme initial rate study
[NAD+]=375, 750, 1500, 3000 uM
[DHP2c]=0, ? uM[NAM]=0, 50, 100 uM
% DMSO = 0%
Time points: 0, 5, 10, 20, 30, 60, 120 min8*2*4*3= 192 reactions----------4 days experiment 1 day data analysis (Feb 03-09, 2016)
Updated schedule (1/19/2016)
DHP2c solubility measurement will continue parallel. For now, 19.5ug/ml=58.1 uM of DHP2c in assay buffer seems to dissolved. Therefore, more test need to be done between 39ug/ml and 19.5ug/ml to test if the solubility is higher than 58uM. (Jan 11, 2016)
Standard curve
%DMSO = 0, 0.001% and 0.01%.[DHP2c] = 0uM, 5uM, 10uM, 25uM, 50uM
3*5*8 = 120 reactions. 2 days (Jan 11 - Jan 12, 2016)
Inter-day variations at 0.001%/0.01%, 0.05%, 0.1%, 0.2%, 0.5%, 1%, 2%, and 5% DMSO
(8*8*2=128 reactions for duplicate and 8*8*3 = 192 reactions for triplicate) (one day for experiments and half day for data analysis) (Jan 13 – Jan 14, 2016)
Finish intra-day/inter-day variation report (Jan 15, 2016)
XG: Attached
Standard curve_complete comparison.pptx
Calibrate the pH probe and measure pH for 100uM DHP2c in assay buffer solution (Jan 15, 2016)
XG: pH = 8.17 at 20.1oC
SI preparation for standard curve (This is a hard task. I just had a outline of it. Please make comments on it. Then I will modify it accordingly. )
XG: Attached
SI preparation for standard curve.docx
@SI draft should proceed concurrently. Emphasize why the SIRTainty assay (other commercial available ones) were not selected for our current study.
@Request COA for different batch of standard stock from Enzo (1/18/2016)
XG: Waiting for response.
@Request standard curve for different lot of standard from Enzo (1/18/2016)
XG: Waiting for response.
EC1.5 measurement with purified protein
[DHP2c] = 0, 10, 25, 50, 75, 100, 150, 200 uM
% DMSO = 0, 0.05%, 0.5%
Time points: 0, 60 min
9*3*2*2=108 reactions----------2 days (Jan 18- Jan19, 2016)
If the 0.05% can do then omit 0.5% one.
Standard curve for EC1.5 calculation
[DHP2c] = 0, 5, 10, 25, 50, 100, 150, 200, 300 uM
% DMSO = 0, 0.05%, 0.5%
9*8*3=216 reactions -------- 3 days (Jan 20-Jan 22, 2016)
@One more set of standard curve for inter-day variation evaluation.
@Try higher [Standard] and test linearity
If the 0.05% can do then omit 0.5% one.
New Enzo enzyme initial rate study
[NAD+]=375, 750, 1500, 3000 uM
[DHP1c]= 0, 50uM
% DMSO =0, 5%
Time points: 0, 5, 10, 20, 30, 60, 120 min
8*2*4 +4*8 = 96 reactions---2 days experiments and 1 day data analysis (Jan 25- Jan 27, 2016)
Purified enzyme initial rate study
[NAD+]=375, 750, 1500, 3000 uM
[DHP2c]=0, ? uM[NAM]=0, 50, 100 uM
% DMSO = 0%
Time points: 0, 5, 10, 20, 30, 60, 120 min
8*2*4*3= 192 reactions----------4 days experiment 1 day data analysis (Jan 28- Feb 03, 2016)
Updated schedule (1/14/2016)
DHP2c solubility measurement will continue parallel. For now, 19.5ug/ml=58.1 uM of DHP2c in assay buffer seems to dissolved. Therefore, more test need to be done between 39ug/ml and 19.5ug/ml to test if the solubility is higher than 58uM. (Jan 11, 2016)
Standard curve
%DMSO = 0, 0.001% and 0.01%.[DHP2c] = 0uM, 5uM, 10uM, 25uM, 50uM
3*5*8 = 120 reactions. 2 days (Jan 11 - Jan 12, 2016)
Inter-day variations at 0.001%/0.01%, 0.05%, 0.1%, 0.2%, 0.5%, 1%, 2%, and 5% DMSO
(8*8*2=128 reactions for duplicate and 8*8*3 = 192 reactions for triplicate) (one day for experiments and half day for data analysis) (Jan 13 – Jan 14, 2016)
Finish inter-day variation report (Jan 15, 2016)
Calibrate the pH probe and measure pH for 100uM DHP2c in assay buffer solution (Jan 15, 2016)
SI preparation for standard curve (Jan 15, 2016)
RC: Ok I see the schedule for these tasks.
MM kinetics_ purified protein (first - just Km, vmax then all if we choose to proceed with initial rate Expts at that concentration).
% DMSO = 0
[NAD+]= 375, 750, 1500, 3000uM
[DHP2c]= 0, 5, 10, 25, 50 uM
Time points: 0, 5, 10, 20, 30, 60, 120 min
4*5*8= 160 ---------3 days (Jan 18- Jan 20, 2016)
RC: See my comments below; I am not sure this is useful, since we will be determining [DHP2c] for paper experiments based on EC1.5 analysis.
Unless you want to do interday analysis (but even that we may be able to do when we get to the paper's initial rate experiments below).
Instead, the enzo enzyme study is important.
Let me know if you have questions/comments.
XG(1/15): You are right. I will move it from the schedule.
EC1.5 measurement with purified protein
[DHP2c] = 0, 5, 10, 25, 50, 100, 150, 200, 300 uM
% DMSO = 0, 0.05%, 0.5%
Time points: 0, 60 min
9*3*2*2=108 reactions----------2 days (Jan 21- Jan22, 2016)
Standard curve for EC1.5 calculation
[DHP2c] = 0, 5, 10, 25, 50, 100, 150, 200, 300 uM
% DMSO = 0.05%, 0.5%
[DHP2c] =100uM at 0% DMSO
9*8*2 + 8=152 reactions -------- 2 days (Jan 25-Jan 26, 2016)
RC: Need to settle plan (time required) for standard curves and reduction of number [DHP2c] at higher DMSO as per comments below if needed, since this was
apparently not accounted for.
XG(1/15): OK. The [DHP2c]s in which the saturation is spotted can be picked for standard curve.
RC: Ok, so you will be selecting concentrations for the standard curves after the EC1.5 experiments. That is reasonable.
Since it appears DHP2c is soluble in buffer below 100uM, we may end up having several concentrations in that range at which we do standard curves.
If I understand correctly, it seems that since you will be reducing the number of DHP2c concentrations for DMSO, this may take less than 2 days.
New Enzo enzyme initial rate study
[NAD+]=375, 750, 1500, 3000 uM
[DHP1c]= 0, 25uM
% DMSO = 5%
Time points: 0, 5, 10, 20, 30, 60, 120 min
8*2*4=64 reactions----------2 days (Jan 27- Jan 28, 2016)
RC: You will be doing 0% DMSO as well, right?
XG: Yes. As your comments listed below, I will do 0DMSO for 0uM DHP1c, and 5% DMSO for 0 and 50uM DHP1c. I will update the schedule after go through all you questions below.
RC: Ok please let me know when you anticipate providing those analyses and when you will finish preliminary discussion with Alok today, so I can be available as needed regarding judgment on assays.
Since you have reduced the planned number of EC1.5 standard curves for FdL, and since these are endpoint rather than mechanistic studies, next week's experiments may not be affected much by this judgment.
Also, the MM experiments with Enzo enzyme and DHP1c planned for the following week can certainly use FdL since that is how we did the biorxiv studies.
The need for many standard curves and more importantly, developer reproducibility issues may create some issues for FdL in the case of mechanistic analyses.
Since the purified enzyme initial rate studies will start near end of month, the HPLC assay may be ready by that time. We may consider aligning schedules for FdL and HPLC if we do both with purified enzyme, and determine
when each will be done.
Purified enzyme initial rate study
[NAD+]=375, 750, 1500, 3000 uM
[DHP2c]=0, ? uM[NAM]=0, 50, 100 uM
% DMSO = 0%
Time points: 0, 5, 10, 20, 30, 60, 120 min
8*2*4*3= 192 reactions----------4 days experiment 1 day data analysis (Jan 29- Feb 05, 2016)
AU: 1-22-2016:
For above reaction which will have 192 samples, HPLC method will take about 38 days plus time to analyse the data.
XG(1/14)
-Since the assay buffer standard curves appear similar to those with 0.01% DMSO (to be verified as priority), we may skip over the MM kinetics for 0.01 or 0.001% DMSO in the schedule below.
XG: The linearity of standard curves in Assay buffer or 0.01% DMSO are similar. Then do you mean, for MM kinetics, only DHP2c in assay buffer will be performed?
-Also, will doing MM kinetics first not be comparable with the later complete initial rate studies including NAM? It will be somewhat redundant in assay buffer. In that case we might just delay this experiment and do all initial rate studies with purified enzyme together. Please comment.
XG: It will be some redundant in assay buffer. For later complete initial rate studies, if we do duplicate at that time, then we may be able to report triplication data.
-We might replace the 0.01% DMSO MM kinetics in the schedule with the planned 0, 5% DMSO MM kinetics with Enzo enzyme for purpose of updating biorxiv. This could come after the EC1.5 experiments with purified enzyme and before the complete initial rate experiments with purified enzyme.
XG: little confusion. Is the MM kinetics in 0.01% DMSO for DHP2c? As mentioned “MM kinetics first - just Km, vmax then all if we choose to proceed with initial rate Expts at that concentration”, the initial MM kinetics experiments designed is for DHP2c at different concentration for just Km and Vmax. Then based on the outcome, we will decide to proceed initial rate experiments at the chosen [DHP2c]s. RC: No the concentration of DHP2c for the purpose of the paper's initial rate data will be based on the outcome of the EC1.5 experiments.
The MM kinetics mentioned here was for the purpose of verifying that the effect of the 0.01% DMSO on the vmax, Km was very small so it does not raise issues with mechanistic analysis or review.However, now that we have DHP2c dissolved in assay buffer, it would not be needed and we could skip to next planned expts (EC1.5 directly).The 0,5% DMSO Enzo enzyme MM kinetics experiments were for another reason: to update biorxiv before someone claims we did not properly account for effects of DMSO (and issues with standard curves).XG(1/15): I got it. I will change the schedule accordingly.RC(1/15): Ok, it seems in that case that we will be able to finish all the EC1.5 experiments and related standard curves next week.
*Important: please note that the Enzo enzyme studies need to be done with DHP1c, not 2c because that was used in biorxiv. For biorxiv, I believe you have already refitted with the new standard curves - please confirm.
XG: Yes.
After the above 0, 5% DMSO MM kinetics study with Enzo enzyme we will see how the DMSO affects initial rates at [NAM]=0. However, we will probably not immediately have time to update all the nonzero [NAM] data in biorxiv.
XG: To make it clear, at 0% DMSO, [DHP1c] will be 0uM. At 5% DMSO, [DHP1c] = 0, 25uM.RC: It needs to be 50uM DHP1c at 5% DMSO since that is what we did in biorxiv. Otherwise, ok.
-Note that in order to properly study the EC1.5 curves; you will need to generate standard curves at each relevant DHP concentration. I assume you are accounting for this in the schedule below. This may be time consuming: please comment.
XG: I did not count on how long will take for extra standard curves will take. I need to know the [DHP2c] range first after Ec1.5 measurement. Then perform the standard curve. For now, we have 0, 5, 10, 25, and 50uM in 0% DMSO on hand. RC: Ok, please account for it so we can decide whether to streamline or not per comments below.XG(1/15): As mentioned above, it will take 2 days to do standard curves under all the conditions. At 0.05% and 0.5% DMSO, we will choose [DHP2c]s where saturation is detected.
For assay buffer, the highest we would be able to go would be 100uM DHP in the EC1.5 study; however, we may see nonlinearity in standard curve close to that.
XG: True. Based on current data, the linearity of standard curve at 50uM DHP2c in assay buffer is OK. I need to test 100uM to confirm.RC: It is good to do 100uM; because we want to verify that we do in fact detect the solubility limit through the nonlinearity of the standard curve. When do you plan to do this?XG(1/15): Will combine with those for EC1.5.RC(1/15): Ok
Since these are endpoint studies, we may be able to efficiently do some of the EC1.5 curves later with HPLC if needed. If it is too time consuming with FdL, we may do only the 0% DMSO (assay buffer) EC1.5 curve for now and others with HPLC later. But we would need to know when Alok can do them.
XG: I agree.RC: Please consult with Alok tomorrow on this and let me know. We may need to prioritize some of these experiments if the FdL standard curves are not reproducible.If needed thereafter we can meet together with him.
Alternatively, we can reduce the number of [DHP2c] for the other DMSO %'s to quickly determine whether we observe saturation.
XG: OK
I did you previously did many such standard curves for DHP 1c EC1.5 study, but not for DHP 2c - please confirm and indicate how long it took.
XG: You are right. I did not do many standard curves for DHP2c EC1.5 study. The EC1.5_DHP2c measurement will be performed first and then the respective standard curves will be scheduled.
-Please let me know when you will switch enzyme batches to the new enzyme from Sudipto within the schedule.
XG: OK.
-Regarding SI on standard curves, we may want to make a note of the reasons we did not use assays like Sirtainty. Based on the notes you provided, do you feel the case can be made as to why these would not be appropriate for initial rate studies?
XG: I can include a paragraph to address the issue you mentioned above. The you can decide if we keep it in the paper or as a draft for responding reviewer’s comments in the future if there is any.RC(1/15): Yes we should do that. This assay-related issue in SI will in any case connect up with our judgment on the reproducibility of the FdL assay/developer protocol mentioned below.
In addition to providing the below as a priority, please also plan to measure the pH of the solutions when the probe comes in on Fri.
XG: The pH probe comes today (1/14/2016). The pH of 100uM DHP2c in assay buffer solution will be measured after the probe is calibrated tomorrow (1/15/2016) sometime.
--Ok, not all the data in assay buffer / 0.01% DMSO are comparable (due to some experiments not being carried out under both conditions), but as I understand the standard curve linearity is similar between the two cases. Please confirm.
XG: The linearity of standard curves in Assay buffer or 0.01% DMSO are similar.
Please include the 0uM DHP 2c in assay buffer standard curve (i.e., the usual standard curve we used without DMSO or DHP) and comment on whether it is valid to compare with these standard curves.
XG: The standard curve in assay buffer (no DMSO and no DHP2c) was included into the table below. The samples were prepared on 1/12/2016 (same day as the rest of samples). However, because of the limit space, the 0 DMSO+0 DHP2c samples were kept in 4oC overnight and run on next day (1/13/2016). The only difference was that the 1X Developer solution was made at different days.
RC: This is difficult to interpret without a direct comparison; see my emails.- At first glance, it appears the nonlinearity at higher [standard] has increased in the experiments this month.This raises some concern about whether there are issues with solubility at lower DMSO %.- However, on closer look, we see that there is a greater nonlinearity (compared to 5% DMSO) also at [DHP]=0uM in 0.01% DMSO.This suggests there may be some issues with interday variability / FdL developer protocol.- There may not be direct evidence that there is an issue with solubility of DHP2c at these concentrations in either assay buffer of 0.01% DMSO.- Please make direct comparison in ppt, and comment on interday variability analysis, etc vis-a-vis report on that topic as well as standard curve SI section writeup tomorrow,consult with Alok on HPLC, and we can discuss immediately thereafter if needed. We need to leave some time for discussion of the interday variability analysis of FdL developer in this context. We need to check whether HPLC will simplify things and save time if there are issues with FdL reproducibility, even if the startup time is a little longer.||
[DHP2c], uM**
| 0% DMSO |
|
| 0 |
487.64 |
| 5 |
464.22 |
| 10 |
381.27 |
| 25 |
331.98 |
| 50 |
297.38 |
--Second, I would like to see a direct comparison (e.g. through combination of ppts) with the standard curves at higher DMSO (where we saw nonlinearity set in above a particular [DHP]).
In those cases, did we also see a difference in linearity with / without the 15uM standard point?
XG: There is a difference in linearity with/without 15uM point (see slide 3 and slide 6).
Standard curve at different [DHP] at different DMSO_1.14.16.pptx
|| || 15uM
| point |
||
| [DHP1c], uM |
Yes |
No |
| 10 |
122.39 |
137.37 |
| 25 |
113.65 |
128.18 |
| 50 |
116.67 |
135.67 |
| 100 |
95.726 |
not linear |
| [DHP2c], uM |
||
| 5 |
174.24 |
184.16 |
| 10 |
162.76 |
186.44 |
| 25 |
158.67 |
176.26 |
--Third, I would like to know how high peptide/standard concentration is relevant in the experimental assays. Do we only operate within the linear range at or below 7.5uM?
XG: In experimental assay, the [peptide/standard] can go as high as 7 uM in control, and ~ 20 uM in addition of DHP1c w 5%DMSO.
XG(1/11):
Based on Dr Raj's latest comments, the updated schedule is listed below
Finished tasks were in gray
- DHP2c solubility measurement will continue parallel. For now, 19.5ug/ml=58.1 uM of DHP2c in assay buffer seems to dissolved. Therefore, more test need to be done between 39ug/ml and 19.5ug/ml to test if the solubility is higher than 58uM. (Jan 11, 2016)
- Standard curve
%DMSO = 0, 0.001% and 0.01%.
[DHP2c] = 0uM, 5uM, 10uM, 25uM, 50uM--------3*5*8 = 120 reactions. 2 days (Jan 11 - Jan 12, 2016)
- Inter-day variations at 0.001%/0.01%, 0.05%, 0.1%, 0.2%, 0.5%, 1%, 2%, and 5% DMSO
- SI preparation for standard curve –half day (Jan 14, 2016)
- MM kinetics first - just Km, vmax then all if we choose to proceed with initial rate Expts at that concentration.% DMSO = 0, 0.001% (0.01% DMSO will be eliminated if the standard curves were linear in assay buffer and 0.001% DMSO)[NAD+]= 375, 750, 1500, 3000uM[DHP2c]= 0, 5, 10, 25 uMTime points: 0, 5, 10, 20, 30, 60, 120 min
4*4*8*3 =384 reactions (Include 0.01% DMSO)---------5 days (Jan 15- Jan 21, 2016)
- EC1.5 measurement[DHP2c] = 0, 5, 10, 25, 50, 100, 150, 200, 300 uM% DMSO = 0, 0.05%, 0.5%Time points: 0, 60 min8*3*2*2=96 reactions----------2 days (Jan 21- Jan22, 2016)
- New Enzo enzyme initial rate study[NAD+]=375, 750, 1500, 3000 uM% DMSO = 0, 5%Time points: 0, 5, 10, 20, 30, 60, 120 min8*2*4=64 reactions----------2 days (Jan 25- Jan 26, 2016)
- Purified enzyme initial rate study[NAD+]=375, 750, 1500, 3000 uM
[NAM]=0, 50, 100 uM% DMSO = 0%Time points: 0, 5, 10, 20, 30, 60, 120 min
8*2*4*3= 192 reactions----------4 days experiment 1 day data analysis (Jan 27- Feb 03, 2016)
XG(1/8): Schedule update
Standard curve 0.01% and 0.001% DMSO with aforementioned [DHP2c]--------2*4*8=64 reactions. one day (Jan 11, 2016)
For current, DHP2c is dissolved in Assay buffer with concentration of 58.1uM. To confirm if DHP2c in assay buffer is greater than 58.1uM, further test is needed. (Jan 11, 2016 - )
Inter-day variations at 0.001%/0.01%, 0.05%, 0.1%, 0.2%, 0.5%, 1%, 2%, and 5% DMSO (8*8*2=128 reactions for duplicate and 8*8*3 = 192 reactions for triplicate) (one day for experiments and half day for data analysis) (Jan 12 – Jan 13, 2016)
SI preparation for standard curve –half day (Jan 13, 2016)
MM kinetics first - just Km, vmax then all if we choose to proceed with initial rate expts at that concentration.
0.01% or 0.001% DMSO
[NAD+]= 375, 750, 1500, 3000uM
[DHP2c]= 0, 5, 10, 25 uM
Time points: 0, 5, 10, 20, 30, 60, 120 min
8*4*5=160 reactions---------4 days (Jan 14- Jan 19, 2016)
EC1.5 measurement
[DHP2c] = 0, 5, 10, 25, 50, 100, 150, 200, 300 uM%
DMSO = 0.05, 0.5, 1
Time points: 0, 60 min
8*3*2*2=96 reactions----------2 days (Jan 20- Jan21, 2016)
New Enzo enzyme initial rate study w and w/o 5% DMSO
[NAD+]=375, 750, 1500, 3000 uM%
DMSO = 0, 5%
Time points: 0, 5, 10, 20, 30, 60, 120 min
8*2*4=64 reactions----------2 days (Jan 22- Jan 25, 2016)
RC (1/7): Updated is required including revised schedule and results to date from solubility study.
XG: -Solubility study is still on (Jan 6, 2016 – Jan 8, 2016)
DHP2c solubility is tested in H2O and Assay buffer.
Every concentration will be prepared and wait for 1 hour to make sure there is no further dissolving.
So far, in H2O, 26ug/ml= 77.5 uM, 19.5 ug/ml = 58.14uM, 46.5uM, 38.8uM, 33.2uM, 29.1uM have tried and need to go lower than that.
In Assay buffer 116.3 uM has tried and so far not dissolved yet, will continue tomorrow for lower concentration
-At the meantime, started to evaluate the intra-day and inter-day variations experiment today, at 0.05%, 0.1%, 0.2%, 0.5%, 1%, 2%, 5% DMSO. Will take 3 days. (Jan7-Jan11, 2016)Will update schedule tomorrow.
RC:
Feedback and necessary updates to schedule:
1) Standard curves
-something seems wrong with standard curve slopes at 0uM dhp and nonzero DMSO - inconsistent for. 1c and 2c.
You show that slope increases at 0 uM DHP for 2c but decreases for 1c.
XG: The slopes at 0uM DHPs with different DMSO in the range 0.05%-5% were listed below. As mentioned the slope increases along %DMSO for DHP2c. There is no clear trend for DHP1c.
| % DMSO |
[DHP1c]=0 uM |
[DHP2c]=0 uM |
| 0.05 |
114.11 |
138.87 |
| 0.1 |
110.4 |
122.45 |
| 0.2 |
105.87 |
132.31 |
| 0.5 |
96.088 |
169.07 |
| 1 |
123.22 |
185.88 |
| 2 |
115.64 |
188.36 |
| 5 |
118.2 |
192.26 |
RC: This does not really answer the question. What do you mean the slope increases for DHP2c but not DHP1c, when both are at 0 uM?So DHPs are not even there and you seem to be running the same experiment in both cases. What are you getting at here? Are you suggesting that the experiment is not reproducible?XG: The aforementioned experiments are at same conditions. The reasons why we did not get similar results are(1) DHP1c and DHP2c samples were run separately not same day.
(2) The developer and Fluor de Lys standard stock were from different lot. The reason different lot was used is because for Fluor de Lys standard stock solution it comes as 30ul per kit and kept in -80oC. To avoid thaw/refrozen cycle, plus only 30ul /tube, normally we use a same reagent for same day experiments and new reagent for different day experiments.
(3) To evaluate this intra-day variation, a duplicate/triplicate experiments need to be performedat same day and different days.
RC: You obviously will need to take this into consideration and provide reliable data on the standard curves. You should schedule accordingly.
2) Solubility
- when you indicate that a compound is soluble or not, what is your protocol? At what concentration do you test?
XG: By eye. We can do future evaluation by using Dynamic Light Scattering (DLS).
-- For aqueous solution
(1) Choose solvents, H2O, 5% NaOH, 5% HCl, and Assay buffer.
(2) Since DHP1c is not soluble in H2O and DHP2c has similar structure, 0.025mg of DHP2c was used which was the value used for DHP1c.
(3) Weigh desired amount (25ug) of DHP2c and then add into 500ul of aforementioned solvents.
(4) Vortex and let it sit at room temperature.
(5) Sometimes it takes very long time to observe if it’s dissolved depending on the solubility.
(6) DHP2c dissolved in 5% NaOH means it has phenol in its structure. To save the sample and time + 5% NaOH will not be the solution used for assay, the future solubility measurement was not performed at this point.
(7) The compound in H2O, assay buffer, and 5% HCl are not soluble for days.
--For organic solution (DMSO is our current interest)
(1) Since Log P value for DHP2c is greater than DHP1c. 5mg/ml is the starting point.RC: This depends on why 5mg/mL was chosen - see below.By P value do you mean octanol water partition coefficient? Latter value is lower for 2cXG: It's Log P.
(2) Weigh desired amount of DHP2c and then add into aforementioned solvents.
(3) Vortex and let it sit at room temperature.
(4) Calculate for solubility value. The test was run up to 70mg/ml. To save the sample and time, the future solubility measurement was not performed, which concluded as solubility of DHP2c in DMSO >70mg/ml.
RC: The issue here is that the statement that DHP2c is insoluble in water may well not be correct, given the predicted log P and DMSO solubility data compared to both resveratrol and honokiol (which are both characterized as being water soluble, and in fact used as drugs in vivo - see Science, Nature Communications, etc) are more favorable for DHP2c. In other words,it may be the case the DHP2c's water solubility is similar to that of some of the well-recognized (and in the case of resveratrol, mechanistically characterized) sirtuin activators.You tested at 50ug/mL, whereas the above two drugs have water solubilities below that? Please provide the molarities of these solutions for resveratrol/honokiol. How much lower is it than 25uM?
XG:
Trans-Resveratrol M.W. = 228.2 g/mol Trans-resveratrol data sheet.pdf
Solubility of trans-Resveratrol inEtOH, DMSO, and dimethyl formamide = ~ 65 mg/ml = 284.8 mM
Solubility of trans-Resveratrol in PBS (pH=7.2) =~100ug/ml = 438.2 uM.
In “Stability and solubility of trans-resveratrol are strongly influenced by pH and temperature” published on European Journal of Pharmaceutics and Biopharmaceutics 93 (2015) 196–204, it was mentioned that UV light causes isomerization of trans-RSV in minutes, pH effects differently on its degradation rate (Trans-RSV was stable in acidic pH because its hydroxyl groups were protected from radical oxidation by positively charged H3O+. On the contrary, trans-RSV was not stable in alkaline conditions. Its degradation followed first order kinetics with reaction rates that increased exponentially in the pH range 6.8–8.0. The degradation profiles above pH 8 were more complex and cannot be described by simple first order kinetics), and increased temperature (such as 37oC) enhances the degradation.
Honokiol M.W. = 266.33 g/molhonokiol data sheet.pdf
Solubility of Honokiol in DMSO = ~33 mg/ml = 123.9 mM
Solubility of Honokiol in 1:4 (ethanol: PBSpH 7.2)= ~0.2mg/ml = 750.95 uMRC: Honokiol is a natural product that has been used in traditional medicines (e.g., Chinese) for a long time.Are there no reports of aqueous solutions/solubility, other than the mention that it is sparingly soluble?XG: Data from Santa Cruz Biotechnology shown Honokiol soluble in DNSO (36 mg/ml), EtOH (<1mg/ml) at 25oC, H2O (>=53mg/ml) at 25oC, caustic alkali, and organic solvents. The differences between Santa Cruz Biotechnology and Cayman Chemical are kind of big, which may due to different synthesize procedure?
DHP2c M.W. = 335.35 g/mol
Solubility of DHP2c in DMSO > 70 mg/ml = 208.7 mM
50 ug/ml = 149.1 uM
RC: So this is a significantly higher concentration than the concentrations of DHP2c used in our kinetic studies. Why would we demand such a high concentration when testing solubility?XG: For DHP1c, it was claimed as water insoluble reagent. Since the similarity of both DHP1c and DHP2c, we did not started at low concentration. At the point we decided what concentration need to be started with, Resveratrol and Honokiolin were not under consideration.
Also, resveratrol was found to have max activation at 100uM - how does this compare to its water solubility?
XG: Solubility of resveratrol in PBS (pH 7.2) is approximately 438.2 uM (100ug/ml). 100uM resveratrol solution should be PBS solution.
And, how did you chooose 25ug in 500uL? Is this an arbitrary concentration for the test of solubility?
XG:25ug in 500ul H2O was used for DHP1c. This is not an arbitrary concentration for the test of solubility.
RC: Used by whom? You? Why was this concentration chosen?XG: Although DHP1c was claimed as water insoluble reagent. I still did try to dissolve DHP1c into assay buffer since there is a pH difference. That is the value I tried. I show you the insoluble solution last time when you were in the lab.
RC: Ok, so the value tried was arbitrary for DHP1c and hence DHP2c as well (and higher than the concentrations at which we did initial rate experiments).
Let's settle these points before proceeding too far with the initial rate experiments (Km,kcat) at low % DMSO
XG: Yes. Will test DHP2c solubility in H2O and assay buffer for lower concentration (<50 ug/ml)
RC: If you continue to encounter issues with solubility, test the pH of the resulting solutions as well and report here.XG: OK
- Please describe the protocol you used to dissolve dhp 2c in assay buffer. Did you account for the possibility that the rate of dissolution may be slow at room temp?
XG: DHP2c firstly dissolved in DMSO to make 50mM stock solution. Then diluted into different concentration using Assay buffer. The calculations is listed below.RC: So your protocol does not allow for the possibility of making a purely aqueous solution. As long as the aqueous/buffer solubility of a compound is less than 50 ug/mL, you always require that there is some DMSO in the solution, even if the desired solution's concentration is < 50 ug/mL (as it is in this case).
XG: The assumption of making buffer/DMSO solution was that DHP2c is not soluble in aqueous solution. The series experiments listed below were aim for finding the lowest DMSO concentration in which DHP2c can dissolve comfortably and with smallest influence of addition of DMSO. The key point here is that we need to find out if DHP2c is soluble in aqueous at low end.
| % DMSO |
[DHP], mM |
DMSO, ul |
DHP 2c, ul |
Assay Buffer |
Total solution volume |
1500 uM NAD+, ul |
| 0.05 |
0 |
0.5 |
0.00 |
969.5 |
1000 |
30 |
| 0.05 |
0.005 |
0.4 |
0.10 |
969.5 |
1000 |
30 |
| 0.05 |
0.01 |
0.3 |
0.20 |
969.5 |
1000 |
30 |
| 0.05 |
0.025 |
0 |
0.50 |
969.5 |
1000 |
30 |
| 0.1 |
0 |
1 |
0.00 |
969.0 |
1000 |
30 |
| 0.1 |
0.005 |
0.9 |
0.10 |
969.0 |
1000 |
30 |
| 0.1 |
0.01 |
0.8 |
0.20 |
969.0 |
1000 |
30 |
| 0.1 |
0.025 |
0.5 |
0.50 |
969.0 |
1000 |
30 |
| 0.2 |
0 |
2 |
0.00 |
968.0 |
1000 |
30 |
| 0.2 |
0.005 |
1.9 |
0.10 |
968.0 |
1000 |
30 |
| 0.2 |
0.01 |
1.8 |
0.20 |
968.0 |
1000 |
30 |
| 0.2 |
0.025 |
1.5 |
0.50 |
968.0 |
1000 |
30 |
| 0.5 |
0 |
5 |
0.00 |
965.0 |
1000 |
30 |
| 0.5 |
0.005 |
4.9 |
0.10 |
965.0 |
1000 |
30 |
| 0.5 |
0.01 |
4.8 |
0.20 |
965.0 |
1000 |
30 |
| 0.5 |
0.025 |
4.5 |
0.50 |
965.0 |
1000 |
30 |
| 1 |
0 |
10 |
0.00 |
960.0 |
1000 |
30 |
| 1 |
0.005 |
9.9 |
0.10 |
960.0 |
1000 |
30 |
| 1 |
0.01 |
9.8 |
0.20 |
960.0 |
1000 |
30 |
| 1 |
0.025 |
9.5 |
0.50 |
960.0 |
1000 |
30 |
| 2 |
0 |
20 |
0.00 |
950.0 |
1000 |
30 |
| 2 |
0.005 |
19.9 |
0.10 |
950.0 |
1000 |
30 |
| 2 |
0.01 |
19.8 |
0.20 |
950.0 |
1000 |
30 |
| 2 |
0.025 |
19.5 |
0.50 |
950.0 |
1000 |
30 |
| 5 |
0 |
50 |
0.00 |
920.0 |
1000 |
30 |
| 5 |
0.005 |
49.9 |
0.10 |
920.0 |
1000 |
30 |
| 5 |
0.01 |
49.8 |
0.20 |
920.0 |
1000 |
30 |
| 5 |
0.025 |
49.5 |
0.50 |
920.0 |
1000 |
30 |
The final solution was used for preparation of different concentrations of standard solutions.
I did not compare the dissolution rate between room temperature and 37oC.
- note that resveratrol has a lower solubility in DMSO (16 mg/mL) than dhp2c (70 mg/mL) and its solubility in water is 3 mg / 100mL (about 0.2 % of its DMSO solubility). Also resveratrols log p is higher (small aqueous partition) than that calculated for dhp 2c.
did you test such low concentrations of 2c in assay buffer before concluding it is insoluble?
XG: The lowest I went for DHP2c was 25ug/500ul = 0.05mg/ml which is greater than 3mg/100ml. I can try 0.03mg/ml and lower.RC: You should go as low as the concentrations we are actually testing in initial rate studies, here 25uM. Otherwise if there is solubility at that concentration and we add DMSO anyway, it would pose a problem.
- are you using method 1 or 2 with dhp 2c?
XG: Method 1 was used for DHP2c which gives lower % DMSO. Method 2 provides constant 5% DMSO.
- note that resveratrol has ec1.5 of 46uM and can achieve 200% max activation, despite having less favorable log P and DMSO solubility than DHP 2c.
guan to check what concentration of resveratrol corresponds to max activation
XG: OK. Double check Sirtris’ 2007 Nature paper “Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes”, ~ 100uM of resveratrol achieved the 200% maximum activation.
guan to check whether any cosolvents were used to achieve max activation
XG: OK. The cosolvents were not mentioned in both article and SI section. Should I email the authors to ask?
- also note honokiol is also less soluble in DMSO than DHP 2c (33 mg/mL) and is said to be sparingly soluble in water. Honokiol actually violates lipinskini rule of 5 with log P > 5
- we can go below EC1.5 with DHP 2c if needed
XG:OK.
XG: Still working on the rest and will update soon.
3) Refitting
- Given that vmax approximately triples in 5% DMSO for Enzo as well (but here for 50uM rather than 25uM DHP 1c) we cannot necessarily claim that the DMSO effect is much smaller in Enzo buffer. May not be worth changing buffer.
- Check whether the extent of activation by 5% DMSO in the endpoint studies is consistent.XG: Still working on it.
- Guan to double check her refitted parameters and to report the relative extents of activation in the endpoint studies.XG:OKRC: In the above two comments I was referring to the fact that in endpoint expts, we saw significantly lower activation by 5% DMSO for Enzo enzyme (compared to in house), and hence we should check if the initial rate experiments (where we saw tripling of vmax) are consistent.
- In any case we did see a comparatively small increase in vmax induced by dhp for purified enzyme and an increase in Km and vmax by 5% DMSO by roughly same factor. - We will check these for enzo enzyme as well; differences may be attributed to purity.XG: OKRC: This is already done, right?
- In initial rate expts, we need 3 sets: no DMSO, DMSO, and DMSO/dhp. Present the results without DMSO in the SI onlyXG: OK
4) Scheduling
- For PNAS priorities are analogous initial rate studies w DHP 2c and 0.05% (or even lower) DMSO. First try even lower % DMSO (eg 0.01% and 0.001%).
XG: Experiments designed
(1) DHP2c concentration(s) need to be determined, 0uM, 5uM, 10uM, 25uM
(2) Standard curve 0.01% and 0.001% DMSO with aforementioned [DHP2c]
--------2*4*8=64 reactions. one day (Jan 5, 2016)
- Do MM kinetics first - just Km, vmax then all if we choose to proceed with initial rate expts at that concentration.
If possible do 3 km,vmax for these concentrations around dec 30-31st
XG: 0.01% or 0.001% DMSO
[NAD+]= 375, 750, 1500, 3000uM
[DHP2c]= 0, 5, 10, 25 uM
Time points: 0, 5, 10, 20, 30, 60, 120 min
8*4*5=160 reactions
---------4 days (Jan 6- Jan 11, 2016)
- Then check extent of activation by DHP 2c relative to low % DMSO control (should be close to ec1.5).
- Then consider adding ec1.5 determination for DHP 2c in presence of 0.05% DMSO and/or 1 or 2 higher concentrations to see if we can reach saturation. Start w a slightly higher concentration and if we can get saturation try lower
XG: EC1.5 measurement
[DHP2c] = 0, 5, 10, 25, 50, 100, 150, 200, 300 uM
% DMSO = 0.05, 0.5, 1
Time points: 0, 60 min
8*3*2*2=96 reactions
----------2 days (Jan 12- Jan13, 2016)
- We may still need to do the 5% DMSO initial rate studies with enzo enzyme as originall scheduled by Guan below if we want to validate v1 of biorxiv. This can come at end after pnas is finished (not on 12/30,31).
XG: New Enzo enzyme initial rate study w and w/o 5% DMSO
[NAD+]=375, 750, 1500, 3000 uM
% DMSO = 0, 5
Time points: 0, 5, 10, 20, 30, 60, 120 min
8*2*4=64 reactions
----------2 days (Jan 14- Jan15, 2016)
5) Miscellaneous
- Guan to provide update on status of, schedule SI prep
XG:
Have obtained most of the data (not include 0.01% and 0.001% DMSO for DHP2c),
the remarks were listed on ppt. Need to gather them and make it logically
smooth flow. Using free time between the experiments.
- Guan to indicate how much enzyme is remaining; how many replicates are possible for planned initial rate studies?
XG: ~550 reactions are left. The planned initial rate studies can be done duplicate not triplicate.
- What if we need to do saturating studies? Use a newly purified batch if needed for initial rate studies? Guan to indicate how much delay. Can use Alok/ Sudipto's enzyme preparations and make enough for all planned expts.
XG: Will use the newly purified batch if necessary from Alok/ Sudipto’s enzyme.
Are you familiar with this commercial assay?
XG: No. The most earliest report was in August 2013, EMD Millipore, the life science division of Germany's Merck KgaA, reports that its new SIRTainty Class III HDAC Assay has been used in a recent study demonstrating a novel mechanism for the activation of SIRT1.
I have been interested in understanding what advantages your coupled continuous assay has over the Hplc based assay we are currently using. Throughput?
XG: As known, two major types of Sirt3 assays were widely used in the past. The difference of which depends on whether fluorophore is included in the reaction. For example, the Sirt3 kit of Enzo Life Sciences includes a fluorophore-linked substrate, which, upon deacetylation, releases fluorophore which can be detected under certain wavelength and therefore generates readout for Sirt3 activity. Another major type of Sirt3 kit, such as the one mentioned by Smith el al, uses a different strategy, in which, instead of using fluorophore, a third reaction was introduced to convert NADH to NAD+ using Glutamate dehydorgenase, the product of which can be detected and serves as the Sirt3 readout (NADH is self-fluorescent at 340nm) . Two major advantages of the continuous assay are (1) no fluorophore induced; (2) the amount of NADH will be recorded and no quenching is needed which means we can take as many time points as we want.
The limitation for HPLC method is not high through put which is not desired for drug screening.
Is the sirtainty assay similar to the coupled continuous assay? I have not yet reviewed it closely .
XG: The sirtainty assay has similar idea as coupled continuous assay. The mechanism behind is listed below
To perform the SIRTainty assay, a sirtuin enzyme, b-NAD, acetylated peptide substrate, test compound, and nicotinamidase enzyme are combined and incubated for 30 minutes. During
this time the acetylated peptide substrate is acted upon by the sirtuin enzyme to produce nicotinamide. In a secondary reaction, the nicotinamidase enzyme converts the nicotinamide
into nicotinic acid and NH3+ (free ammonia). To generate a signal for readout, a proprietary developer reagent is added and the signal is read using a fluorescent plate reader.
Advantages of this assay
(1) No fluorophore induced to peptide substrate
(2) Other peptide substrate can be tested using this assay
(3) Good for measuring sirtuin activity as well as for screening of activators and inhibitors of sirtuin enzymes
(1) High through put
Disadvantages of this assay
(1) It’s not clear what the chemistry behind the developer reagent like Fluor de Lys assay.
(2) Need to be quenched. In other word, the readout need to be developed by adding developer reagent.
(3) Also not clear if the reaction will be stopped by adding the developer. For different time points, it’s probably not desirable for kinetic study.
If this assay is available and we didn't use it, we may want to explain why.
XG: OK.
----------------------------------------------------------------------------------------------
In addition to my previous email about sirtainty assay, you should compare the assay 1 below and also consider what is value of number 2 below, and update.
XG: References were attached.
A Novel Continuous Assay for the Deacylase Sirtuin 5 and Other Deacetylases_2015_steegborn.pdf
A FRET-based assay for screening SIRT5 specific modulators_2015_Yan.pdf
Fluorescence assay of SIRT protein deacetylases using an acetylated peptide substrate and a secondary trypsin reaction_2004_Marcott.pdf
Method 1 and 2 are using same principle. Method 2 was published 5 method earlier. The principle behind this method was based on the 2004 Analytical Biochemistry paper below.
---- Fluorescence assay of SIRT protein deacetylases using an acetylated peptide substrate and a secondary trypsin reaction. Analytical Biochemistry 332 (2004) 90–99
The authors synthesized a SIRT peptide substrate based on a segment of the p53 sequence containing the acetylated lysine and then modified this peptide to devise a fluorogenic substrate suitable for screening. The polypeptide was rendered insensitive to trypsin proteolysis by replacing all arginine and underivatized lysine residues with serine; thus, the substrate contains one acetylated lysine residue that is readily deacetylated by SIRT2. A fluorogenic substrate for SIRTs was devised by attaching fluorescent tetramethylrhodamine-6-carboxylic acid (6-TAMRA) and quenching (QSY-7) groups on opposite sides of this trypsin-resistant sequence. No increase in fluorescence is observed on incubation of the substrate with SIRT1 or SIRT2. The deacetylated product, but not the substrate, is readily cleaved by trypsin, and the fluorescence increase after the addition of trypsin is a measure of the deacetylase activity.
Substrate peptide for SIRT2: Ac-EE-K(QSY-7)-GQSTSSHSK(Ac)-L-Nle-STEG-K(6-TAMRA)-EE- NH2.
- A Novel Continuous Assay for the Deacylase Sirtuin 5 and Other Deacetylases. J Med Chem. 2015 Sep 24;58(18):7217-23
2. A FRET-based assay for screening SIRT5 specific modulators. Bioorg Med Chem Lett. 2015 Apr 15;25(8):1671-4.
A fluorescence resonance energy transfer (FRET)-based assay where a donor dye and a quencher dye are connected to an acetyl peptide substrate.A FRET effect will occur in this acetylated peptide, which will cause the fluorescence of a donor dye quenched by a quencher dye. The deacetylation of sirtuins followed by trypsin digestion disrupts the FRET signal and thus releases the fluorescence from a donor dye.
Assay principle
Substrate peptide used for SIRT5
Ex/Em = 340 nm / 490nm.
Remarks for this type of assay: Specific peptide substrates is needed.
XG: Schedule update
12.30.2015 – 12.31.2015 New Enzo enzyme initial rate study w and w/o 5% DMSO
---[NAD+]=375, 750, 1500, 3000 uM
---% DMSO = 0, 5
---Time points: 0, 5, 10, 20, 30, 60, 120 min
01.04.2016 Concentration measurement of Combined batch I+II+III
prepare the SI section for standard curves
01.05.2016 – 01.06.2016 Combined batch I+II+III, measurement for Km/ Vmax, in assay buffer/5% DMSO/5% DMSO+25uM DHP1c (2 days)
Standard curve at different [DHP1c]_12.21.2015.pptx
New fitting using new standard curve.xlsx
EC1.5 calculation using standard curve at different [DHP1c]
EC1.5 Calculation using standard curve at different [DHP1c]_12.02.15.pptx
RC: Ok, not sure why the AFU values are so much higher for Enzo (don't have complete info on how the samples were prepared);but, it is interesting to note that EC1.5 is very similar for both, despite greater activation of in-house by 5% DMSO that we saw in the ppt below.This may have implications for the mechanism of action of DHP vs DMSO (related to the comments in my recent email). Will elaborate later.
XG(12.16)
--Enzo enzyme and in house enzyme are in same dialysis buffer (stock solution), which includes
25mM Tris (pH=7.5), 100mM NaCl, 5mM DTT, and 10% Glycerol.
--The compositions of reaction buffers for Enzo and in house enzymes system are listed below
| Enzo |
In house |
|
| Tris/Cl |
50mM |
42.5mM |
| pH |
8.0 |
7.5-8.0 |
| NaCl |
137mM |
125.9mM |
| KCl |
2.7mM |
1.89mM |
| MgCl2 |
1mM |
0.7mM |
| BSA |
1mg/ml |
0.7mg/ml |
| Dithiothreitol |
0 mM |
1.5mM |
| Glycerol |
0% |
3% |
The reasons of causing the differences are(1) Enzo enzyme has better activity but 70% purity. Less amount of Enzo stock solution will be added into the reaction buffer system.
(2) In house enzyme has less activity but >90% purity. More stock solution is needed which changes the composition of the reaction buffer.
- How do you determine the amount of enzyme / U you want in the reaction? Is it arbitrary? What is the difference in the amounts of stock soln needed in each case?- In the future, given that we know the activity of in house enzyme, could we adjust dialysis buffer so the final buffer compositions of the Enzo and in house reactions are the same?
XG(12.15):DHP1c/DMSO related data
Calculation of DMSO final concentration for Method 1 and Method 2.xlsx
Batch I and II Vmax Km.xlsx
Dependence of enzyme activity on DMSO concentration_12.14.2015.pptx
XG (12.15): Schedule update
12.16.2015: identify the source of apparent inconsistency between endpoint and initial rate data. DHP2c solubility in H2O and DMSO
12.17.2015-12.18.2015 and 12.21.2015:
Standard curve
%DMSO: 0, 0.05%, 0.1%, 0.2%, 0.5%, 1%, 2%, 5%
[DHP1c]=0, 5, 10, 25 uM
[DHP2c]=0, 5, 10, 25 uM
[Standard] = 0, 0.234375, 0.46875, 0.9375, 1.875, 3.75, 7.5, 15
Prepare the report and refit biorxiv data12.22.2015 Concentration measurement of Combined batch I+II+IIIprepare the SI section for standard curves
12.30.2015 – 12.31.2015 Combined batch I+II+III, measurement for Km/ Vmax, in assay buffer/5% DMSO/5% DMSO+25uM DHP1c (2 days)
XG(12.14): Km in the absence of DHP in assay buffer and in 5% DMSO.xlsx. The standard curves used for batch I and II were included in the file. No clear why the inconsistency occur.
purposed next experiments
(1)Standard curve, 0% DMSO, 5% DMSO and 10% DMSO with same developer (1day)
(2)Combined batch I+II+III, measurement for Km/ Vmax,in assay buffer/5% DMSO/5% DMSO+25uM DHP1c (2 days)
XG(12.14): Schedule update
[NAD+] = 375, 750, 1500, 3000uM
[DHP1c]=0, 50 uM
[NAM]= 0, 50, 100uM, and higher concentration
12.04.2015 SIRT1 EC150 measurement if the reagent is delivered to the lab.
12.07.2015 [NAM] = 0, 50 uM; DHP= 0 uM, 50uM, [NAD+]=375, 750uM.
12.08.2015 [NAM] = 0, 50 uM; DHP= 0 uM, 50uM, [NAD+]=750, 1500uM
12.09.2015 [NAM] = 0, 50 uM; DHP= 0 uM, 50uM, [NAD+]=1500, 3000uM
12.10.2015 [NAM] = 0, 50 uM; DHP= 0 uM, 50uM, [NAD+]=3000, 375uM
12.11.2015 Review and analyze data
12.14.2015 and 12.15.2015 determination of higher NAM concentration. Concentrated NAM solution may change the ionic strength/pH of the reaction solution which might affect the (1) enzyme performance (2) developer chemistry. Will do a titration of [NAM] in range 200-500uM.
12.16.2015 [NAM] = 100uM, higher concentration; DHP= 0, 50uM, [NAD+]=375, 750uM
12.17.2015 [NAM] = 100uM, higher concentration; DHP= 0, 50uM, [NAD+]=750, 1500uM
12.18.2015 [NAM] = 100uM, higher concentration; DHP= 0, 50uM, [NAD+]=1500, 3000uM
12.21.2015 [NAM] = 100uM, higher concentration; DHP= 0, 50uM, [NAD+]=3000, 375uM
12.22.2015 Review and analyze data
RC: The following was sent by email on Sat 12/12. The answer to the first question is important at this time.
1) I think I am missing something in this data. You report a Km of 2000 in the absence of DHP. Previously, didn't you report a Km closer to 1000? Please explain. Please elaborate in your answer so we can have the most efficient communication.
XG: The previous experiments in the absence of DHP was done in assay buffer. The 5% DMSO was in the presence of DHP experiments. Therefore, to keep consistency, 5% DMSO was included in the current experiments (in the absence of DHP). Also the 5% DMSO standard curve was used for calculation.
RC: I thought you had mentioned that 5% DMSO had been used in the past for control experiments for consistency.
I need to consider this carefully. Please send me the earlier kinetic results (esp Km, vmax) in assay buffer for no DHP (you can just list them here if you want, alongside the new ones).
Please also post the xls you sent last week here.
Km in the absence of DHP in assay buffer and in 5% DMSO.xlsx
25uM DHP1c_12.11.2015.xlsx
RC: Since there was DMSO, my earlier statement about regarding standard curves does not apply - they are important. I see you applied the 5% DMSO standard curve.
Different concentration of DHP1c in 5% DMSO_11.30.15.pptx
I would like to review the earlier data on DMSO effects on activity as well. As I recall in the endpoint experiments you saw apparent activation in the presence of lower DMSO concentrations. Unless I am mistaken, you had applied the DMSO standard curves here.
Dependence of enzyme activity on DMSO concentration_11.23.2015.pptx
2) It would be useful to have the data regarding higher NAM concentration. We would like to do at least one higher concentration, though the results will determine how high we can go without compromising the assay. Are you doing initial rate experiments or endpoint for this? When you do the higher [NAM], it would be helpful to have a Dixon-type plot at a particular NAD+ (slope of 1/v vs [NAM]). Please advise.
XG: The higher NAM concentration will be done using endpoint method. Few time points will be measured to make sure the higher [NAM] will not compromise the assay. It is a good idea to have a Dixon-type plot.
RC: I would need to know the answer to the issue of higher NAM concentration by EOD Tues.
XG: I will start the assay 12.14.2015 and send you report EOD Tues.
RC: Since we are not doing saturating experiments, we may consider doing an additional duplicate experiment if time permits and sufficiently high NAM is not achievable (based on your alternate schedule provided, but perhaps extending into a following week to get triplicate data).
XG: OK. When we get the higher [NAM] results, I will update the schedule accordingly.
3) I will have some further comments about model fitting once this experimental series is finished, so please consult with me shortly before the series is done and before final fitting.
XG: OK.
RC: Please let me know your days in office for December in advance as well. I will then advise on next experiments thereafter. They may involve either a few more control experiments or another DHP (we should have 2 other DHPs in lab by then).
XG: Sherry talked to me early Nov about my schedule. I still have 6.5 vacation days for this year. Because of the tight experimental schedule, she said that I can carry my vacation days to 2016. Therefore I will not take all my vacation days off this year. I need few days to visit my previous MS degree advisor’s family. I have a very good relationship with them. They treated me as their own daughter. They send gifts to my children every Christmas. Normally we will call each other during holiday season since they are at Toronto. Unfortunately Prof. Pan (my advisor) got brain cancer two years ago and passed away. Last week, I got to know that Prof. Feng (his wife) has issue with her heart. She wants to see me and my family ASAP. I will take three days off for the trip, which are 12/23, 12/28, and 12/29. Thank you for your understanding.
RC: That's fine of course, I just wanted to know the schedule for planning purposes.
RC: Let me know when DHP 2c arrives.
XG: OK.
Please recall that you will also need to be preparing the SI section for standard curves.
XG: Will do between the experiments
XG(11.06): Urea method update
Urea method update_11.06.15.pptx
XG(11.02): Schedule update
11.03.2015-11.06.2015 purify protein 1mM IPTG/8M Urea treatment Batch 3 and Batch 4
11.09.2015-11.10.2015 Prepare new DHP1c stock solution and measurement of EC1.5 for DHP1c using
newly purified protein, saturating DHP 1c concentration needs to be decided.
11.11.2014 [NAM] = 0, 50 uM; DHP= 0 uM, saturating, [NAD+]=375, 750uM.
11.12.2015 [NAM] = 0, 50 uM; DHP= 0 uM, saturating, [NAD+]=750, 1500uM
11.13.2015 [NAM] = 0, 50 uM; DHP= 0 uM, saturating, [NAD+]=1500, 3000uM
11.16.2015 [NAM] = 0, 50 uM; DHP= 0 uM, saturating, [NAD+]=3000, 375uM
11.17.2015 Review and analyze the first set of [I] concentration
11.18.2015 [NAM] = 50, 100uM; DHP= 0 uM, saturating, [NAD+]=375, 750uM
11.19.2015 [NAM] = 50, 100uM; DHP= 0, saturating, [NAD+]=750, 1500uM
11.20.2015 [NAM] = 50, 100uM; DHP= 0, saturating, [NAD+]=1500, 3000uM
11.23.2015 [NAM] = 50, 100uM; DHP= 0, saturating, [NAD+]=3000, 375uM
11.24.2015 Review and analyze the second set of [I] concentration
11.25.2015 [NAM] = 100 uM, higher concentration; DHP= 0 uM, saturating, [NAD+]=375, 750uM
11.30.2015 [NAM] = 100 uM, higher concentration; DHP= 0, saturating, [NAD+]=750, 1500uM
12.01.2015 [NAM] = 100 uM, higher concentration; DHP= 0, saturating, [NAD+]=1500, 3000uM
12.02.2015 [NAM] =100 uM, higher concentration; DHP= 0, saturating, [NAD+]=3000, 375uM
12.03.2015 – 12.04.2015 Put all three set of data together and analyze, model fitting.
RC: We may omit the higher NAM concentration depending on our assessment of the time required to complete paper (which will add further days to schedule above).Will advise at appropriate time.
XG(10.28): Urea method update
Urea method update_10.28.15.pptx
RC (10-23):Regarding schedule below for this week, please confirm that we will be finishing the following today:
-batch to batch reproducibility comparison
XG(10.23): Large scale purification (two batches) was performed this week. The total amount, concentration, and specific activity, of purified protein are comparable between these two batches.
RC: Good. It sounds like the yield is reduced from the previous week, when you anticipated having enough from one batch?XG(10.23): Since we do not have enough protein, I start culturing cells again, the OD 600 is monitored and then IPTG induction will be performed. The culture will be grown over night, then harvest Saturday morning. Monday and Tuesday will purify another two batches. Concentration/ specific activity will be measured on Wednesday. I would expect to have enough protein by then.
RC:I thought the proposal below was to purify two batches this week. The question was whether there would be enough in one batch for all experiments and if not, we could combine the two purified this week?XG(10.23): The total purified protein we got this week can do 397 reactions. We were planning to receive proteins can complete 800 reactions.
RC : Do you mean 397 reaction from two batches, rather than the expected 800 (400 per batch)?XG(10.23): That is from two batches. 216 from batch I and 181 from batch II.
So you got about 50% of last week's yield?XG(10.23): I got more protein with less specific activities.
RC: I am not understanding the significant differences between this week and the last couple of weeks.I believe there were multiple batches previously done with urea, and several of them had > 0.40 specific activity.As I recall, previously, 8M urea gave > 0.40 specific activity twice, and 6M urea gave 0.45 specific activity.The good thing is that there is quite a minimal batch to batch variation - one can't expect much less variation than this.XG(10.23): Previous weeks, I did 1 bottle (200mL) per day. The sample proceeding time was ~ 45 mins. Now 4 bottles need more than 2 hours.
Please revise proposed schedule accordingly.
XG(10.23):
10.26.2015-10.28.2015 purify protein 1mM IPTG/8M Urea treatment
10.29.2015-10.30.2015 steady state parameter estimates (Km,vmax) for control
11.02.2015-11.03.2015 Prepare new DHP1c stock solution and measurement of EC1.5 for DHP1c using
newly purified protein, saturating DHP 1c concentration needs to be decided.
11.04.2015 [NAM] = 0, 50 uM; DHP= 0 uM, saturating, [NAD+]=375, 750uM.
11.05.2015 [NAM] = 0, 50 uM; DHP= 0 uM, saturating, [NAD+]=750, 1500uM
11.06.2015 [NAM] = 0, 50 uM; DHP= 0 uM, saturating, [NAD+]=1500, 3000uM
11.09.2015 [NAM] = 0, 50 uM; DHP= 0 uM, saturating, [NAD+]=3000, 375uM
11.10.2015 Review and analyze the first set of [I] concentration
11.11.2015 [NAM] = 50, 100uM; DHP= 0 uM, saturating, [NAD+]=375, 750uM
11.12.2015 [NAM] = 50, 100uM; DHP= 0, saturating, [NAD+]=750, 1500uM
11.13.2015 [NAM] = 50, 100uM; DHP= 0, saturating, [NAD+]=1500, 3000uM
11.16.2015 [NAM] = 50, 100uM; DHP= 0, saturating, [NAD+]=3000, 375uM
11.17.2015 Review and analyze the second set of [I] concentration
11.18.2015 [NAM] = 100 uM, higher concentration; DHP= 0 uM, saturating, [NAD+]=375, 750uM
11.19.2015 [NAM] = 100 uM, higher concentration; DHP= 0, saturating, [NAD+]=750, 1500uM
11.20.2015 [NAM] = 100 uM, higher concentration; DHP= 0, saturating, [NAD+]=1500, 3000uM
11.23.2015 [NAM] = 100 uM, higher concentration; DHP= 0, saturating, [NAD+]=3000, 375uM
11.24.2015 – 11.25.2015 Put all three set of data together and analyze, model fitting.
As I understand you will be doing kinetic analysis concurrently.
XG(10.23): I see. The kinetic analysis is based on purified protein. I obtained the final product from o/n dialysis this morning, and then characterized the concentration, calculated the specific activity. Based on the specific activity data, steady-state parameter estimation experiment will be designed and performed.RC: I meant concurrently with the subsequent purifications - as stated in the schedule above.
I assume you are starting kinetic data analysis today?XG(10.23): As scheduled below "10.19. 2015 -10.23. 2015-- Grow bigger amount of cells (8x200ml each), then harvest, lysis, treat with Urea, column purification, dialysis, characterization (concentration, specific activity). Two batches will be done. The variation will be compared from batch to batch." Cell growth took two days; harvest and purified and dialysis took two days (4 bottles per day); protein characterization took one day; steady state parameter estimates (Km,vmax) for control will take another 2 days. Actually, as mentioned in .ppt, proceeding 4 bottles a day which took a lot time before loading onto column, might be one of the reasons that specific activity dropped down.
RC: What is meant by proceeding 4 bottles a day? Processing more bottles than before? If so, why would this reduce activity - some of it goes bad before column loading?XG(10.23): You are right. We only proceed one bottle (200mL) per day before. Because we scale up, we were planning to load samples proceeded from 4 bottles (4x200mL) to one column for one batch. These 4 bottles have to be proceeded in one day for purification. That is why we do 4 bottles a day. Possible reasons of decreasing activity- For each 200mL culture, it takes quite a while to suspend and proceed for Urea treatment. Proceeding time was too long for 4-fold of cell (800ml) in one day. Homogenizer is needed for speed up these particular steps. - Loading 4X samples on 1 single column does not provide 4x amount of protein- Some loss from FlowThrough indicated the binding was not sufficient- Purity was decreased because of the insufficient washRC: Ok. Please propose how you will change protocol to deal with this next week (expanding as needed on points made in ppt; does the revised schedule above reflect these changes?)
XG(10.23): Proceed 3 bottles every day and purify with separate column. Then dialysis them together. The revised schedule reflect these changes.
I will review the ppt in the meantime.
-determination of whether we have enough yield from a single batch for plans below or whether two batches would need to be combinedXG(10.23): Analysis shown that we do not have enough yield from a single batch. Please see the attached .ppt for details. Urea method update_10.23.15.pptx
-steady state parameter estimates (Km,vmax) for control (?)
XG(10.23): Need 2 days to provide the estimate.
Regarding the schedule for the following weeks, please comment on the following:
- 2 interday duplicates of every NAM concentration except 0,higher (i.e., for 50,100uM) are planned.
this appears to provide triplicate data for 50,100uM NAM. (if the NAMs were not repeated, we would have done 100uM, higher in second week; so there is one additional week).
is there another reason for this besides obtaining this triplicate data, like always repeating a previous day's expt? as you mentioned, one overlapping experiment is planned for each successive day.
XG(10.23): Overlapping experiment by each successive day can provide quick feedback and indicate if the results from previous day are good;
XG: Urea method update 10.16.15
Urea method update_10.16.15.pptx
RC (10-16):-Please compare the yield between 8M urea this week and 8M last week. Comment on whether the protocolmodifications this week aimed at reducing the amount of protein loss in pellet were successful (it appears yieldincreased).XG: It's hard to conclude if the yield is improved from gel picture. However, this week's 8M urea provide more final protein, which may due to (1) 1 mM IPTG induction we have more protein to start with; (2) increasing the incubation time on ice (from 5 min to 30 min), (3) putting sample on shaker during incubation.
-Please indicate whether there was some purity improvement at 8M since last week (95% pure this week).XG: About the same.
-Please post original papers that described the urea method.cloning express and purification of human truncated SIRT1_2012.pdf
RC: I don't think this is one of the original papers that described the method itself. Please provide a original references (not for sirtuins) where the method was first described, or the most detailed reference available on the methods.XG(10.19): The original paper for Urea method was addressed as protein refolding method. please check the paper below:
Clark_Current Opinion Biotechnol. 9, 157-163.pdf
Based on this, people modified the method for their applications.
-Is the number of batches required for a single set of initial rate experiments referring to with or without intraday duplication?XG: without intraday duplication
How many required with intraday duplication, and for both 50uM and saturating DHP?XG: 1 batch
Regarding schedule for next steps:-For now please operate under assumption that we will moving on directly with initial rate studies using urea method purification starting next week-Please describe in detail how we will scale up expression/purification so we can get all the above experiments done with a single batch. Assume intraday duplication. Do we need to schedule technical visit for FPLC?
XG: We would grow more cells and proceed and purify through same column which consider to one batch. The maximum capacity for proceeding cells for one batch is 4 X 200ml cell culture. Ideally we will obtain ~ 4-fold of amount of protein, which can provide total amount of protein for ~ 800 reactions. We need to grow such amount of cells at same time then purify them. Then after characterization, we will have rough idea, if the amount of reactions for one batch reaches our expectation (~ 800reactions).
RC (10/19): What if it doesn't? Will combine two batches?XG(10.19): Yes, will combine the two batches.
800 reactions is sufficient for intraday duplication (2x) for both 50uM and saturating DHP?XG(10.19): Yes, for [NAM]= 0, 50, 100uM. If another higher [NAM] is needed, then another 400 reactions are needed (include 2x).
RC: Need to consider this in advance.
-Provide the schedule for the next few weeks, day by day, including purification and initial rate studiesThis includes description of the [NAD],[NAM] you will do each day. (You don't have to repeat every detail for saturating in the protocol description).-Assume the initial rate studies may include one higher [NAM]XG:
10.19. 2015 -10.23. 2015 Grow bigger amount of cells (8x200ml each), then harvest, lysis, treat with Urea, column purification, dialysis, characterization (concentration, specific activity). Two batches will be done. The variation will be compared from batch to batch.
RC (10/19): Agreed that there should be a batch to batch comparison. Please specify details of how the comparison will be done. Comment on the add'l time required to compare vmax and Km for the two batches.XG(10.19): The comparison between the two batches will be divided into steps:-- the physic-chemical characters: concentration, total amount protein, and specific activity -- one day
-- Control experiment --- [NAM]=0uM, [DHP]=0uM, [NAD+]= 375, 750, 1500, 3000uM --- using two different batch of purified enzyme----2 days
If the urea method for both batches is consistent, the initial rate studies can be turned on. We will call FPLC technical visit after we finish the paper.
10.26.2015-10.27.2015 Prepare new DHP1c stock solution and measurement of EC1.5 for DHP1c using newly purified protein, saturating DHP 1c concentration needs to be decided.
10.28.2015[NAM] = 0, 50 uM; DHP= 0 uM, saturating, [NAD+]=375, 750uM.
10.29.2015 [NAM] = 0, 50 uM; DHP= 0 uM, saturating, [NAD+]=750, 1500uM
10.30.2015 [NAM] = 0, 50 uM; DHP= 0 uM, saturating, [NAD+]=1500, 3000uM
11.02.2015 [NAM] = 0, 50 uM; DHP= 0 uM, saturating, [NAD+]=3000, 375uM
11.03.2015 Review and analyze the first set of [I] concentration
11.04.2015 [NAM] = 0, 100uM; DHP= 0 uM, saturating, [NAD+]=375, 750uM
11.05.2015 [NAM] = 0, 100uM; DHP= 0, saturating, [NAD+]=750, 1500uM
11.06.2015 [NAM] = 0, 100uM; DHP= 0, saturating, [NAD+]=1500, 3000uM
11.09.2015 [NAM] = 0, 100uM; DHP= 0, saturating, [NAD+]=3000, 375uM
11.10.2015 Review and analyze the second set of [I] concentration
11.11.2015 [NAM] = 0 uM, higher concentration; DHP= 0 uM, saturating, [NAD+]=375, 750uM
11.12.2015 [NAM] = 0 uM, higher concentration; DHP= 0, saturating, [NAD+]=750, 1500uM
11.13.2015 [NAM] = 0 uM, higher concentration; DHP= 0, saturating, [NAD+]=1500, 3000uM
11.16.2015 [NAM] = 0 uM, higher concentration; DHP= 0, saturating, [NAD+]=3000, 375uM
11.17.2015 – 11.18.2015 Put all three set of data together and analyze, model fitting.
RC (10/19): The above includes intraday duplication (2x) on each day? (By combining batches, is there any problem with doing 3x intraday duplication?)
XG(10.19): The above does not includes intraday duplication. I was thinking to do inter-day duplicates instead. Because the current schedule has overlapping one [NAD+] at different day, which make more sense instead of doing same [NAD+] again. It will be 100 reactions a day, therefore, no room for the intraday duplication
RC: Do you mean the plate wells will all be occupied every day in the plan above, and hence that interday would be just as easy to do as intraday?And would the time required by 2x that required without duplication?XG(10.19): The plate will be occupied every day in the plan above. Same [NAD+] will provide intraday duplicate and different [NAD+] will provide interday duplicate. The time required will be doubled without duplication.
It actually appears you are doing interday duplication (not intraday) abov, since you are repeating [NAD+] on different days. Please comment.XG(10.19): Yes.
It also appears you are limited to 2 [NAM] a day, for reasons that are not entirely clear, and we end up repeating [NAM]=0 three times.XG(10.19): Good point. 2 [NAM] are the maximum can do per day.However, we can do [NAM]= 50, 100uM for 11.04.2015 - 11.09.2015;[NAM]= 100uM, saturating for 11.11.2015 - 11.16.2015.
RC: Not clear on this. saturating NAM is not of interest. Please reconsider the schedule modification.Did you mean 0,50 uM NAM; then 100 uM, higher concentration NAM?XG: I mean higher concentration NAM.
-Please comment on how much faster the purification could be done with Alok's involvement so we can decide on his schedule for Arctic Express and this in parallel.RC: Please try to post the above today at least in draft form.
XG: Under current exp. condition/schedule, Alok’s involvement may not speed up the purification process. But he will be available to help whenever it’s needed. We only have one set of magnetic stirring system for dialysis, which require overnight occupation; therefore, the purification process is limited.
XG: Protocol 10.14.15Protocol 10.14.15.docx
RC (6-17): Please prepare a tabular schedule on dropbox, like Ping and Alok, for the tasks below and update accordingly. Please include purification tasks on the schedule of tasks in between the assay tasks where relevant.
Summary of 5.29.15 meeting
RC: May need to file experimental provisional patent by Oct due to priority date of publication of PLOS paper (RC to check this). Patent appl would prescribe exptl screening methods for identification of potential activators of multiple sirtuins, not through allosteric activation mechanism of SIRT1.XG: The PLOS ONE online publishing date is Sep. 15, 2014. The patent will mostly focus on the new screening method for identification of potential activators of multiple sirtuins based on the experimental results.RC: Guan to check patent Sirtuin activators and activation assays (mostly SIRT1), which describes some reaction mechanisms for modulation and targeted diseasesXG: WO2011130595A2_Sirtuin activators and activation assaysSIRTUIN ACTIVATORS AND ACTIVATION ASSAYS_WO2011130595A2.pdfUS2015011830A1_Methods and compositions for activation of sirtuins with annexin A1 peptides US20150111830A1_methods and compositions for activation of sirtuins with annexin A1 peptides_2015.pdfThe patents listed above are those the assay methods were mentioned in the patent.Need to look closely to check1) How they describe the methods (format) in a patent2) How they claim the activators based on activation assay3) Please add the necessary points you are interestedRC: Experimental emphasis for paper 2 will be on C pocket binding affinities vis-à-vis associated models for obtaining binding affinity estimates from kinetic data. This should be done in a timely fashion following isoNAM and possibly one DHP inhibitor characterization.XG: The experiments considering being involved in this matter:
1) [isoNAM]=0, 10 mM
[NAM] = 500 uM
[NAD+]= 375, 750, 1500, 3000 uM
RC (6-17): First complete the 500 uM NAM experiment with Enzo enzyme. Is this enzyme from the same lot as that used for lower [NAM] (I believe it is; it needs to be to be comparable).Then do controls (0 isoNAM) and 10 mM isoNAM for all [NAM]'s, using the in-house purified enzyme. According to your email, this should be possible with 100 mL purification scale. How long will the latter take (including the initial 4x100mL stock preparation)?
The experiments should start next week.
2) DHP inhibitor (DHP 1b) mechanistic experimentThe condition of inhibitor experiment will keep the same as what was used in activator experiment[DHP 1b]= 0, 50 uM[NAM] = 0, 50, 100 uM[NAD+] = 100, 375, 750, 1500, 3000uM3) The repeat experiment using in house purified proteinMay only repeat one DHP molecule[DHP 1b/DHP1c]= 50 uM[NAM] = 0 uM[NAD+] = 100, 375, 750, 1500, 3000uM
RC (6-17): This would require another 100 mL scale purification. This repeat could eventually be done for DHP 1c.
We should discuss the results using the Enzo enzyme for DHP 1b first before doing the repeat.
I believe the DHP inhibitor (1b) would use a different Enzo lot than the activator, but that is ok for purposes of patent.
The 1b experiment should be scheduled after the isoNAM experiments. Please estimate in the schedule when these will be done.
There are group of patents about Dihydropyridine on Sirtuins (Patents_Dihydropyridine on sirtuins is attached)Patents_Dihydropyridine on Sirtuins.pdf. I may need to be reviewed latter to understand how to use DHP 1b and DHP 1c into our patent application as an example.RC: However, XG to indicate how long base exchange expts would take (which can provide some relevant info needed); if possible in shorter time than C pocket binding affinity studies, then we may consider doing those instead for 2nd paper.XG: Based on Dr. Sauve’s group published work (Biochemistry 2013), Base-exchange experimentsü [carbonyl-14C]nicotinamide working concentrations: 0, 10, 20, 30, 45, 60, 80, 90, 125, 250, 360, 600 μCi/molü 10ul aliquots at different time points:0, 30, 60, 90, and 120 minü Each aliquot will be separated using HPLC ü The peak for NAD+ will be collected and the radiation will be countedIt will take 5-7 weeks to finish the experiments from method modification to results analysis.RC (6-17): We may opt to do these base exchange experiments on SIRT3 following the assay of one or more additional DHPs (starting with inhibitor 1b, see below).
RC: XG should do one comparison between enzo and in-house purified enzyme eg w isonam to help determine whether we need to redo more expts for our papers schedule. Current exptl data (and possibly more) can be used for provisional patent on workflow. isoNAM expts may need to be finished before we hold another group mtg.XG: The repeat experiment using in house purified protein
[isoNAM]=0, 10 mM
[NAM] = 100 uM
[NAD+]= 375, 750, 1500, 3000 uM
RC: Meet to discuss specific activity and esp activity for 10 mL prep when ready. Determine what scale necessary and what time required to make our own enzyme for a complete initial rate study of a particular inhibitor/activator. Then consider whether purified enzyme can be used for continuing dhp studies (incl dhp inhibitor) or whether we should use commercial enzyme for those.XG:ü Both SIRT3 proteins w/o His-tag show activity. The comparison is calculatedSIRTuin purification_5.29.2015.pptx. The specific activity for both proteins will be measured. ü Meantime the 4X100ml culture will grow in the lab and IPTG induction will be performed. The expressed SIRT3 cell culture will be centrifuged and pellet will be stored into -80oC for the future use. ü Enzo protein will be used for DHP studies. However, the repeat should be performed in one DHP modulator at 50uM. RC (6/17): As noted above and discussed, we will first to dhp inhibitor using commercial enzyme since its first application will be in patent, and we want rapid results.We will later do/redo certain dhp experiments with in-house purified enzyme for publication.
Please estimate how much faster it will be to do each dhp initial rate study (i.e., for each distinct dhp compound) using commercial vs in-house purified enzyme (i.e., compare the time required for initial rate measurements alone to that of initial rate measurements + purification). This will allow us to determine how many DHPs we could assay in a few months' time.
RC: Appears we will need to use purified enzyme for both C pocket binding affinity studies, wt characterization (which would be controls) and --isoNAM-- for papers. Still, since we plan to compare enzo and in-house purified enzyme for isoNAM, we can proceed w both isoNAM and control (esp since we always repeat control for each new modulator) before larger scale purification.XG: OK.Could do DHP inhibitor study for patent w commercial enzyme (after isoNAM with commercial and isoNAM with pure).From patent perspective, base exchange expts are of lower priority.RC: LIA - the C pocket binding affinity correlation with the IC50 data. IC50 data were obtained using both endpoint and initial rate methods. What are the differences between these two methods?XG: For IC50 measurement, we got similar results from either initial rate method or endpoint method. In most of the publications, the IC50 values were reported using initial rate method. In the past, we used endpoint method to explore the good range of ligand concentration first. Then use initial rate method to get results for the purpose of publication.
For one ligand, IC50 measurement reactions were performed in the presence of one fixed NAD+ concentration, and 6 different concentrations of C-Pocket-binding Ligand. The initial rates were measured at (0, 10, 20, 30, 60 min). Those are 30 reactions, another 2 background check, total are 32 reactions. For a 96-well plate, 3 ligands can be tested out at the same time.
Convert IC50 to Ki can be done by using online tool (http://botdb.abcc.ncifcrf.gov/toxin/kiConverter.jsp).
RC (6-17): Please provide the estimated time for assay of 6 ligands for publication using this method and in-house purified enzyme. I would like to compare to the time required for base exchange expts (5-7 wks).After I receive this info, we will schedule the C pocket studies and the base exchange experiments in order or priority.
RC: Is the unit Enzo used universal?
XG: No. Since we would like to compare Enzo protein to in house purified protein, we will use Enzo definition.
ü In general, one U is defined as the amount of the enzyme that produces a certain amount of enzymatic activity, that is, the amount that catalyzes the conversion of 1 micro mole of substrate per minute. The conditions also have to be specified: one usually takes a temperature of 25°C and substrate concentration that yield the maximal substrate conversion rate.
ü Enzo SIRT3: One unit will deacetylate 1 pmol/min of Fluorde Lys substrate at 37oC using 500uM NAD+ and 500uM Fluor de Lys substrate.
XG(5-18): SIRTUIN purification progress is attached.
In brief,
- SIRT3 expression in 100ml scale is good.
- AKTA first HisTrap HP column run has completed.
- Fraction evaluation indicated (1) wash volume can be reduced from 20 to 10 CV; (2) Elution can be reduced to 5 CV.
- His-tag cleavage and dialysis will be performed
- AKTA second HisTrap HP column run will be run
- Purified SIRT3 will be test on activity assay.
XG:
- His-tag cleavage and dialysis will be performed (5/19/2015)
- AKTA second HisTrap HP column run will be run (5/20/2015)
- Evaluation of his-tag cleavage efficiency and fraction content (5/21/15)
- Purified SIRT3 will be test on activity assay (5/22, 5/26)
Will review.RC: -Regarding the posted plasmid sequence maps, were these generated using PlasMapper based on the sequencing data from Origene?
XG: Yes.
-I am not able to find the proposed plasmid sequences from Origene that I thought had been posted in Dec 2014 prior to ordering. I had reviewed/approved them at that time. Please let me know where they can be found.XG: please see attached word files.
CW103308.docxCW103309.docx
-What is the difference between run 1 and 2 in the double digestion gel?XG: Run I and 2 are two separate runs of inoculate cells, culture, miniprep, which indicate the method can provide reproduced results.
-Have you analyzed the double digestion gel data vis-a-vis the maps? If so, did you get the fragment lengths expected?XG: Yes. Please check the attached pdf file for details.SIRT1.2 digestion5.22.2015.pdf
In brief,
- The plasmids of Sirt1 and 2 failed to generate the appropriate proteins. When the restriction digestion was performed, the sequences provided by OriGene don't agree with the results of the restriction enzymatic analysis.
- Next step: (1)Request OriGene analyze SIRT1 and SIRT2 plasmids, and send us both the enzymatic diagnostis and sequencing results. (2)If the problem is observed, the new plasmids need to be made without extra charge.
- Solution: (1) Email has been sent to request both the enzymatic diagnostic from OriGene. (2)OriGene has set up a free replacement (SO#SR031647) to get another vial of these two custom clones. They will do the same digestion and fully sequence the insert genes for these two clones. This replacement should take 2~3 weeks.
-Both expression level and purity appear good for the eluted fractions (pre-His tag cleavage? (we had previously verified expression level (more crudely) prior to running the column)XG: Yes. Pre-His tag cleavage.
-Please provide the protein gels from the post-His tag cleavage for expression level / purity verification when ready. Same wash and elution volumes used?XG: Reduced Wash (from 20cv to 10cv) and elution volume (from 10 cv to 5 cv). For the second AKTA purification, the target protein(without his-tag )will be firstly washed off and collected. The fraction from elution is cleaved his-tag.
-You will be doing activity assays comparing purified protein from the pre- and post-His tag cleavage at the same time. Have these been started?XG: Yes. On the way.
When will you do quantification of purified protein from the 100mL scale for the purpose of specific activity estimates? We will be comparing specific activity of the pre- and post-His tag cleavage samples.XG: The specific activity of the pre- and post-His tag cleavage samples will be tested next week.
RC (5-15): Please post some of your recent results / sequence maps discussed with RC on wiki and continue to do so as work progresses.Schedule: It appears you should have activity measurements finished by third wk of May. We would then be ready to be proceed with either purification scale up or isoNAM kinetic studies. We will also have sequence validation completed on SIRT1 and 2 by roughly that time.
RC: Thanks. Regarding the protocols, are you following those from the papers listed verbatim, or are you making any changes? If so, which changes?Since the protocols differ somewhat for the three enzymes, are you using three different protocols in parallel as you prepare the enzymes for purification (you indicated you would be working on protein expression simultaneously for the three enzymes).
I will review and then possibly stop by the lab to see the status of the work in progress.XG(4/17/15): I am following the protocols listed below. It is possible to have further modification along the progress of the experiments. I will mark the changes as it needed. Currently for sequencing and quick screening if the SIRT1/2/3 can be induced and expressed by IPTG, the method is the same. Different SIRTUIN may require different IPTG concentration and induction time. After the quick screening, we will know and do them differently.
The purification of SIRT1/2/3 will be performed one by one on AKTA.
XG (4/16/2015)
SIRT1/2/3 protein expression and purification are the current focus. Sequencing will be the first step to confirm that the plasmids from OriGene are right ones. The sequences are listed below:
SIRT1
UniProt ID: Q96EB6
Length: 747
Mass (Da): 81,681
SIRT2
UniProt ID: Q8IXJ6
Length: 389
Mass (Da):43,182
SIRT3
UniProt ID: Q9NTG7
Length: 399
Mass (Da): 43,573
Then IPTG induction condition needs to be figured out for the maximum protein expression at certain IPTG concentration. Protein purification s will be next step and human SIRT3 will be the first one to be purified. The protocols for SIRT1/2/3 expression and purification are listed as following. It is possible to have further modification along the progress of the experiments.
M. Pan, H. Yuan, M. Brent, E.C. Ding, and R.Marmorstein. THE JOURNAL OF BIOLOGICAL CHEMISTRY VOL. 287 (2012) 2468–2476.
- A plasmid expressing full-length human SIRT1 was a generous gift from David Sinclair (Harvard Medical School). DNA fragments encoding various SIRT1 constructs (residues 214–583, 160–583, 160–665, and 584–665) were cloned into the first multiple cloning site of the pETDuet- 1 vector containing the gene for yeast SMT3, a ubiquitin like protein of the SUMO family.
- BL21-CodonPlus (DE3 RIL) cells harboring the SIRT1 expression constructs were grown at 37 °C in LB medium and induced at log phase with 0.5 mM isopropyl -D-thiogalactopyranoside at 18 °C overnight.
- The cells were harvested and lysed by sonication in a sonication buffer containing 50 mM Tris, pH 7.5, 200 mM NaCl, 5 mM imidazole, 10 mM -mercaptoethanol and 5% glycerol with the addition of 0.1 mg/ml PMSF.
- The cell lysate was centrifuged to remove cell debris, and the protein supernatant was loaded onto a nickel-nitrilotriacetic acid-agarose column and washed extensively with sonication buffer supplemented with 30 mM imidazole, before eluting the protein with a 30–600 mM imidazole gradient in sonication buffer.
- The His-SUMO tag was removed by incubating the SIRT1 protein with ULP1 (ubiquitin-like-specific protease 1) at 4 °C overnight and then passing the overnight incubation solution through a fresh nickel-nitrilotriacetic acid column equilibrated in sonication buffer plus 30 mM imidazole.
- The NaCl concentration of the collected flow through was lowered to 100 mM by diluting with sonication buffer containing 50 mM NaCl and loading the solution on a HiTrap Q ion exchange column.
- SIRT1 protein elution was carried out with a gradient of 100–800 mM NaCl in sonication buffer.
- Peak SIRT1 protein fractions were further purified on Superdex-200 in a buffer containing 20mM HEPES, pH 7.5, 100 mM NaCl, 5% glycerol, and 10 mM DTT to yield 90% pure SIRT1 protein (as judged by SDS-PAGE).
- SIRT1 protein was concentrated to 10 mg/ml using a micro concentrator and either used immediately or stored at -80 °C after flash freezing prior to use.
- A 6-liter purification typically yielded 1–3 mg of purified protein depending on the specific SIRT1 protein construct.
Human SIRT2 constructs were expressed in Escherichia coli BL21(DE3) strains (Novagen). The cells were grown in minimal media with 100mg/L ampicillin and 4 mM Zn(Ac)2 at 37 °C. When the OD600 reached 0.8–1, 40 mM Zn(Ac)2 was added along with 1mM IPTG.
- The cells were induced at room temperature and harvested after 7 h of induction.
- Soluble protein was purified by nickel-affinity chromatography. Three grams of cell paste was lysed through a microfludizer in 40mL buffer A (20mM Tris–HCl, pH 8.0, containing 200mM NaCl, 10mM imidazole, and 2mM - mercaptoethanol) plus 0.1% v/v Triton X-100 and 1 tablet of complete, EDTA-free protease inhibitor (Roche) to inhibit any protease activities.
- The lysate was centrifuged at 19,500g for 40min to remove the insoluble portion. The clarified supernatant was loaded onto a nickel ProBond Resin (Invitrogen) column.
- After washing with 20 column volumes of buffer A and 10 column volumes of buffer A plus 20mM imidazole and buffer A plus 50mM imidazole, the protein was eluted with buffer A plus 300mM imidazole.
- The Trx-fusion protein was cleaved at 4 °C with 0.2 units of biotinylated-thrombin (Novagen) per mg of protein for 15 h while dialyzing into a buffer containing 50mM Tris (pH 7.5), 100mM NaCl, 5mM imidazole, and 2mM - mercaptoethanol.
- The sample was then incubated with streptavidin agarose to remove the protease, and the fusion protein was removed by passing through a second nickel column.
- SirT2 protein was eluted with 300mM imidazole. The final buffer for SirT2 studies was 50mM Tris (pH 7.5), 100mM NaCl, 5% glycerol, and 5mM DTT.
Crystal Structures of Human SIRT3 Displaying Substrate-induced Conformational Changes. JBC. 284,(2009) 24394–24405
Human SIRT3-(118–399) was cloned into a modified pET21b vector (Novagen) between BamHI and XhoI, which places expression under the control of the T7-lacO promoter. The protein was expressed in E. coli BL21-Gold(DE3) cells (Stratagene) as an N-terminal fusion to a hexahistidine affinity tag with integrated TEV protease site.
- A single colony was inoculated in LB media containing 100 g/ml ampicillin at 37 °C, 250 rpm until the A600 reached 0.3. The culture was then transferred to 18 °C, 250 rpm until the A600 reached 0.6–0.8.
- Isopropyl 1-thio--D-galactopyranoside was added to a final concentration of 0.3 mM, and expression was continued at 18 °C, 160 rpm overnight.
- Cells were collected by centrifugation, and the pellet was resuspended in lysis buffer (200 mM NaCl, 5% glycerol, 5 mM 2-mercaptoethanol, and 25mM HEPES-NaOH, pH 7.5) and sonicated to open the cells.
- Supernatant was separated from cell debris by centrifugation at 10,000xg for 40 min at 4 °C and loaded onto a Ni-NTA column (Qiagen) that equilibrated with the buffer containing 200 mM NaCl, 5% glycerol, 5 mM 2-mercaptoethanol, 20 mM imidazole, and 25 mM HEPES-NaOH, pH 7.5.
- The column was washed with 5 column volumes of the buffer containing 200mM NaCl, 5% glycerol, 5mM 2-mercaptoethanol, 50 mM imidazole, and 25 mM HEPES-NaOH, pH 7.5, and eluted with the buffer containing 200 mM NaCl, 5% glycerol, 5 mM 2-mercaptoethanol, 250 mM imidazole, and 25 mM HEPES-NaOH, pH 7.5.
- The eluted protein was dialyzed in lysis buffer and digested with TEV protease (Invitrogen) to remove the N-terminal His tag at 4 °C overnight.
- The protein was loaded on a second Ni-NTA column equilibrated with lysis buffer. The untagged protein was eluted by the buffer containing 200 mM NaCl, 5% glycerol, 5 mM 2-mercaptoethanol, 5 mM imidazole, and 25 mM HEPES-NaOH, pH 7.5. The purified protein was dialyzed against the dialyzing buffer containing 200mM NaCl, 5 mM 2-mercaptoethanol, and 20mM Tris-HCl, pH 8.0, and concentrated.
- The protein was further purified by a S200 column (GE Healthcare) to 95% purity as assessed by SDS-PAGE analysis stained by Coomassie Brilliant Blue R-250 and concentrated to 10–15 mg/ml in the dialyzing buffer. SeMet protein was expressed inM9media (in-house) and purified by the same method described as above.
XG(1/5/15)
First Priority
- Data analysis SIRT3_isoNAM 0, 100 uM NAM (1/5/15)
- Modifiy schematics in Temp_New_Fig_Slides. Slide 4 is being redrawn as slide 5. The labeling in slide 4 (competitive inhibition of deacetylation, etc) can be used to label the corresponding arrows in the new slide 5. (1/6/15)
- Will include Denu 2007 paper measuring the NAM cleavage rate constant by rapid quench methods into the RATE CONSTANT FIGURE and simplify it_condense into one figure that contains all the relevant annotated information regarding the known rate constants. All notations will be explained by the Figure caption. (1/7/15)
- Rate limiting step for certain sirtuins, SIRT3, SIRT1 Sir2 (1/8/15)
- Mutants and activity effects, including which mutants were studied computationally (Yingkai Zhang’s paper…) (1/8/15)
- Experimental data for small molecules with C pocket binding for LIA (provide standard errors). Looking for FdL assay first. If there is not enough, will test them in lab (1/9/15)
- Patent techniques for structural based assay-AMPK modulator_Screening methods for AMP-activated protein kinase modulators: a patent review.Kim J, Shin J, Ha J.Expert Opin Ther Pat. 2014 Dec 23:1-17.
- Mechanistic study on another DHP class inhibitory molecule (experimental test)
XG(11/12): Draft a response letter to Enzo to address (1) inpurity issue (2) proper labeling on SDS-PAGE gel picture (3) quote for the cost of enzyme with high purity ..
XG(11/17): Done. Waiting for the Quote.
XG(11/12): PLOS One data analysis for different NAD+ concentrations
XG(11/17): The PLOS data has been analyzed at different NAD+ concentration under different NAM concentration. The Excel files (PLOS ONE DATA FITTING_SIRT3_various NAD and NAM concentration and PLOS ONE DATA FITTING_SIRT1_various NAD and NAM concentration) have been uploaded in dropbox PMC-AT Research/Xiangying/
RC: Does this include the data from the previous analysis with all NAD and different NAM?
XG(11/18): Yes.For SIRT3, 5/4/3 NAD concentrations with 4/3/2 NAM concentrations were analyzed. For SIRT1, 4/3 NAD concentrations with 3/2 NAM concentrations were analyzed.
RC(11/20) : For a), examples include the Steegborn paper (2013) that compares NAM IC50 for several different sirtuins. This paper also talks about c) NAM inhibition study on Sir2Af2. For b), some examples include statements that product release is rate limiting for some Sir2's, information you have already presented in your qm/mm slides showing which step of deacetylation is rate limiting for that enzyme vis-a-vis experimental kcat estimation, etc. I have already read these statements in some papers, and you have prepared a preliminary slide on the rate constants previously. This needs to be updated with papers like the one I mentioned by Denu on NAM cleavage rate measurement with rapid quench. These are essential to our papers. Since we have already downloaded and read many of the relevant papers, it should be possible to make discernable progress rapidly even if the analysis is not exhaustive. You should present what you have for review early on, since as with the case below, much of the info may not be relevant to our needs. Also, there may not be much info available for most of the sirtuins.
XG(11/21):
*Some of the aforementioned tasks "....statements in some papers, and you have prepared a preliminary slide on the rate constants previously. This needs to be updated with papers like the one I mentioned by Denu on NAM cleavage rate measurement with rapid quench. These are essential to our papers." have been addressed in .ppt file named as "Sirtuin Chemical Mechanisms_11.21.2014" which located in Dropbox/Sirtuin Chemical Mechanisms_11.21.2014 folder along with the related references.
*Mutants on HST2, Sir2Af2, and hSIRT3 with references were added in Excel table "SIRTUIN_mutants" in dropbox PMC-AT Research/Xiangying/". Will keep updating.
*IC50(NAM) overview is uploaded in dropbox/PMC-AT Research/Xiangying.
*Also make a table with all known mutants and activity effects, including which mutants were studied computationally
XG: As mentioned, I spent 2.5 day to make the table. However it does not still include everything. I need more time to finish that too.
RC: As noted you should omit anything that does not deal with in vitro kinetics/mechanism (which appears to save up to 90% of the work).
XG(11/12): Get quote for SIRT1, 2, and 3 plasmid from Origene
XG(11/14): Have answered their questions and wait for the Quote.
XG(11/12): Literature work
(1) maximum inhibition by NAM for all these sirtuins
(2) All known info on rate constants of all these sirtuins in the same table including which step is rate limiting if known
(3) Also make a table with all known mutants and activity effects, including which mutants were studied computationally
XG(11/14): An exce file: SIRTUIN_mutants has been uploaded in dropbox PMC-AT Research/Xiangying/
(4) NAM inhibition study on Sir2Af2
RC(11-14): plan an isoNAM/NAM series of initial rate experiments as discussed (analogous to DHP/NAM). Here you should choose a single isoNAM concentration where we see activation in presence of NAM. This [isoNAM] will be fixed while [NAM] is varied.Let me know when the planning is complete.
XG(10/17):
*One set experiment of [NAM]=100 uM at 0 and 50uM DHP 1c (10/20-10/24), Data analysis (10/27)
*An endpoint verification experiment with DHP 1b and 1b derivative for SIRT3 and SIRT1 (10/28-10/29).
RC (10/15): Next experiments to schedule:
a) an additional [NAM], at 0 and 50 uM DHP
b) a single day endpoint verification experiment with ethyl phenyl DHP derivative that was found to be an inhibitor in JMC paper
XG(9/12) From 9/12 group meeting
1) Send order request to Risa for purchasing DHP-1b: take ~ two weeks from date of PO;
2) Start Different time point assay on SIRT3 (one [NAD+], and one [modulator]);
3) DHP-1c activity test at 1 uM on SIRT1 (EC150_SIRT1 = 1uM);
4) Dosage dependent assay to test how high [DHP 1c] can go for SIRT3 (EC150_SIRT3=50uM);
5) EC150 of DHP 1b derivative for SIRT1 and SIRT3;
6) Continue 2nd manuscript preparation;
5) Will calculate NAM concentration to decide what [NAM] need to be used for relative inhibition assay.
XG 8/11 wiki post _ initial rates for [NAM]=0, various [NAD] in uM/min for SIRT3
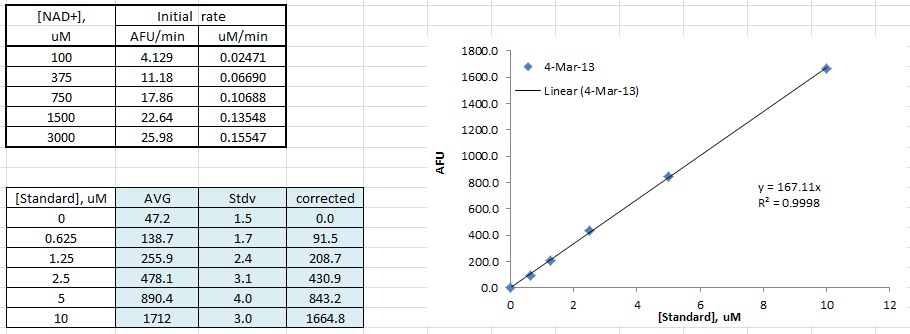
XG(8/25):From 8/22/14 group meeting
*Consider to use TCan for long term solution
*Calculate dosage of radioactive reagent
*Find out the procedure for license application
*Cost for continuous assay vs. Fluor-de-Lys kit
*Need to discuss the NAM concentration during the endpoint assays in JMC paper. Please remind Dr Raj at the appropriate time.
RC (8/7): Over the next month or so (until the continuous assay is finished) you should spend 2/3 of your time as priority finishing the continuous assay and
1/3 of time on the 2nd paper outlining.
Details:
a) given a 2/3 time commitment, please provide/confirm the estimated time for continuous assay completion including validation of the method (e.g. by comparison to FdL results).
XG(8/7):
Large Scale purification of protein PNCA, protein characterization, and activity measurement -----3 weeks.
Validation of the continuous assay----- 2-3 weeks.
b) 2nd paper: points below should start being incorporated as bullet points into appropriate sections of paper draft provided. If any questions, ask. This includes points that map rate constants previously
determined experimentally to our diagram, differences between base exchange rate constant determination method of Sauve and ours - assumptions he made, etc. Bibliography and citations should be started.
This will give me a better idea of length. A major focus of this paper is presentation of the model for sirtuin reaction kinetics, which can be used for either mechanism-based inhibitor and activator design and which was not presented in 1st paper. We will assume PCB as journal for now. Based on the length estimate, we will decide whether to move the derepression activation sections to a separate paper.
c) 2nd paper: some relevant sentences from 1st paper intro (including references, e.g. to NAD+ regulation of sirtuins) should be added where indicated in outline
d) 2nd paper: should add point to intro regarding how allosteric activators work only with SIRT1 and also only with certain substrates with hydrophobic +1/+6 residues. Provide some info on how many SIRT1 substrates do not have this feature, as motivation for other approaches.
e) 2nd paper: we should indicate early on that the paper deals with both mechanism-based inhibitor and activator design. PNAS paper on Ex-527 should be referenced here.
f) Lower priority: after some of above finished, we will discuss what is required to run the cell bio assays for upregulation and downregulation of sirtuin activity in our lab.
g) For relative inhibition section, more detail on the relative inhibition assay is needed. See below. Then add the latest data and protocol to the appropriate section. Some changes to our assays may be needed in the future to account for generation of NAM during reaction.
Prior to further studies on dihydropyridines, we will calculate the [NAM] over time in the endpoint activity assays. Will let you know when.
h) RC will review the list of leads identified through similarity search, especially the substructures used, prior to experiments with those leads. Assuming confirmation, experiments will start with inhibitor leads. These results may be presented in a third paper.
-Please provide lead time for delivery of the molecules identified through similarity search.
XG(8/7): It depends on the company we order from. In general, the chemicals will be shipped out within 10 business days after the order is confirmed.
-Please indicate which substructures were used to generate each lead.
XG(8/11): The substructures are added in the Table.
Similarity search results.docx
-Please provide an estimate of time required for IC50 determinations for each lead using continuous assay. Hits will be passed to kinetic characterization (initial deacetylation rate) experiments.
XG(8/7):
Solubility test (in H2O or DMSO in common or specific solvent)---3 lead/day;
Optimization of range of the [inhibitor]---need 2-3 runs (low, medium, and high [ ]);
Background signal adjustment if compound has color---3 lead/day.
A 96-well plate can be run on TECAN or Fluoroskan for up to 4 compounds ---2 plates/day.
Data analysis will be performed from every run to optimize the final IC50---2 runs/day.
i) Please verify whether any complexes may be amenable to ITC binding affinity measurement (follow up on your group mtg task on this).
XG(8/7): We discussed the possibility of using ITC to bring in solid binding affinity data before. If the modulator potent enough, ITC can be applied (MST was used in PNAS paper for Ex527:NAD/substrate/ Apo-enzyme).
For accurate measurement, the comfortable zone will be 1nM - 100uM.
Time: One run for couple of hours. Then system cool down. For each binding pair, if everything goes smoothly, one would need at least two samples (1) with small molecule, (2) without small molecule_ reference. The concentrations of protein and/or small molecules need to be adjusted for best S/N, which would take more time. Typically people normally plan for 2- day experiment for each binding pair.
RC(8/11): Please indicate whether ITC would be possible for, e.g., dihydropyridines assuming that the Kd is <= 50 uM.
XG(8/11): Yes. ITC can be applied for such measurements. It will be really nice to provide those values for purposes of paper publication and patent application. Since the requirement of bigger amount of protein, we need to make SIRTUIN enzymes in house. As mentioned before, the enzymes(SIRT1/SIRT3) to be used in continuous assay are purchased and cost-consuming. Sooner or later, we need to make SIRTUIN enzyme in house. Because the similarity of the experision and purification procedure, we can make two or three of interested SIRTUINs at once. Roughly, it will take 3-4 month to get them on hand.
j) regarding base exchange assay, we need info on: price of custom-labeled reagents (e.g. NAM), whether reagent vendors can ship to us without any validation of our ability to handle such reagents, price of LSC, whether LSC can be purchased by any lab, confirmation that no local approval is needed for us to use such reagents.
XG(8/11):
PerkinElmer (http://www.perkinelmer.com/Catalog/Product/ID/NEC831010UC)
Sigma-Aldrich
![[carbonyl-14C]NAM_Sigma.jpg [carbonyl-14C]NAM_Sigma.jpg](files/%5Bcarbonyl-14C%5DNAM_Sigma.jpg)
PMC-AT Email at 10/29/2013 9:31 AM
Specific license application for the use of radioactive material in NJ.
Thank you for contacting us about the use of radioactive material in New Jersey. The exempt quantity of C-14, is 100 microcuries, so you will need a license to possess the 34.5 millicuries of material for the experiments. You can access the application under “Forms” and “Specific License” at: www.agreementstate.nj.gov
You would be applying for a “Research & Development – Other” type of license, Program Code 03620, Billing Code of 3M, annual fee of $4285. This fee would accompany your application and would cover the current fiscal year of July 1, 2013 through June 30, 2014.
You could terminate the license at any time ($200 fee). Keep in mind that next fiscal year billing will happen in the summer of 2014.
Please let me know if you have questions – there are instructions with the application. You should submit the check list that is mentioned in the instructions along with the Application Form 313.
Catherine L. Biel
Radiation Physicist
NJDEP Bureau of Environmental Radiation
Radioactive Materials Program
Mail Code 25-01
P.O. Box 420
Trenton, NJ 08625-0420
Office: (609) 984-5663
Fax: (609) 633-2210
Email: [email protected]
Physical Location:
25 Arctic Parkway
Ewing, NJ 08638
Website: www.agreementstate.nj.gov
PMC-AT Email at 10/30/2013 10:43 AM
Waste pickup
Sam Suevo
Operations Manager- Philadelphia Pa
Veolia ES Technical Solutions, L.L.C.
Office: 215-537-7342
Fax: 215-533-6799
Cell: 215-416-5260
Email: [email protected]
Website: http://www.Veoliaes.com/
k) 2nd paper: In PMC AT Research folder, there is a Hubbard 2014 review on sirtuin modulators that discusses inhibitor applications. Please add notes from Hubbard review on SIRT1 modulators (2014) regarding applications of inhibitors as bullet pts to paper draft. There is also a review from 2011 there.
l) Please look up any/all patents on dihydropyridines as drugs - there appear to be many used as calcium channel blockers.
Also please verify that no other other work has been done with these on sirtuins.
XG(8/12):
(1) 1460 dihydropyridines related patents were found and 26 patents were found for SIRTUIN + dihydropyridine
using Patentdocs.
(2) 120,000 dihydropyridines related patents were found and 374 patents were found for SIRTUIN + dihyropyridine (including dulplicates---one title with different modification at different time point) using Google Patent Search.
Example from (2) Patents_Dihydropyridine on Sirtuins.pdf.
RC (8/6): Please see paper attached and buy dihydropyridine derivative 1c, which we discussed, for purpose of repeating the activation study herein and relative inhibition study in presence of NAM (SIRT3 then SIRT1). I assume none of the other molecules herein are available. Please let me know estimated delivery date.
dihydropyridine_activators_mammalian_sirtuins.pdf
XG(8/7): Reagent order request has been sent to Risa. The estimated delivery date will be posted when it's available.
RC(8/7): Please confirm that this is the only derivative available.
XG(8/7): Yes. 1c is the only derivative available. The estimated delivery date will be around 8/21/14.
XG(8/18): Double checked with Risa, and Catalogue #: BML-GR359-0005; Sales order #: SO-0337644; Estimated time for delivery: 08/21/14.
RC (8/18): Ok, please provide a suggested timetable of experiments with this molecule including the initial validation of the results from the paper and then kinetic studies investigating the degree of competition with NAD and peptide.
Some of these experiments could start with FdL (given that the paper also used FdL) while completing the continuous assay preparation. Depending on when the experiments would start, we could plan to discuss the [NAM] produced during the endpoint assay around that time.
RC (8-18): Please check whether the same company can custom synthesize the phenyl derivative 1b.
XG(8/18): Vitas M Labs has the following molecule:
Should I contact Vitas M Labs since they provide similar molecule? Should I order the above molecule (one -CH3 difference)?
RC (8-18): Yes; I believe this is the methyl rather than ethyl ester? I assume the paper did not study this molecule? Please indicate the price then order; also, you should ask them if they can make molecules like 1b for us.
XG(8/18): Quote requst has been sent. Waiting for the response and will update when the info become available.
XG(8/19): Quote is listed below. Have sent to Risa to set an order (10 mg).
XG(819): Still waiting for info of customized compound 1b(left below).
Before running any further relative inhibition assays, I need further clarity on the results of the isoNAM assays. Please update the data in the 2nd paper with the latest results from the isoNAM relative inhibition assays. Was SIRT1 activated? We need to resolve any issues with the assay including reproducibility before publishing in 2nd paper. Are you able to report a EC1.5 (concentration required to boost activity by 50%) based on this data?
Regarding buildup of NAM during reaction: Based on the isoNAM time series data used to generate the initial rate plots for isoNAM inhibition (no NAM), you should have data on the activity of SIRT1 in the presence of e.g. 1mM isoNAM (or whatever concentration gave maximum activation in relative inhibition experiments) and the absence of NAM over the course of ~ 1 hr. Please indicate how that compares to the activity in the absence of isoNAM. This is the closest analog we have to the results reported by Mai for dihydropyridines. I assume the activity will be lower, but I would like to see the numbers.
RC(8/11): Please provide some of the initial rates for [NAM]=0, various [NAD] in uM/min for SIRT3
XG(8/11):

RC (8/6): Before proceeding to answer my new questions below posted today, please indicate approximately how long it will take you to complete each of the tasks, so we may develop a schedule.
XG(8/1/14): Remaining tasks/questions
(1) you mentioned that ”Previous studies have focused on the complete steady-state kinetic mechanism (a) the inherent differences in catalytic efficiency and substrate preference among Sir2 enzymes (ySir2, yHST2, and SIRT2) for various histone peptides; (b) the overall kinetic mechanism; (c) Resolve the individual chemical steps of the Sir2 reaction.”specifically in terms of the rate constants of the reaction mechanism, what does mean? In what way were each of the individual chemical steps resolved? Subsequently, you mention that rapid quench was required to determine particular reaction rates.
XG(8/4): Previous studies have focused on resolving the steady-state kinetic mechanism. In terms of the rate constants, some of the individual step(s) have been measured experimentally or computationally.
(A) Substrate Binding: Using equilibrium dialysis ([3H]AcH3), the binding of NAD+ to HST2 and the binding of acetylated substrate to HST2 were measured. The results supported that the acetylated peptide is first to bind with a Kd of 150 uM. No NAD+ binding was detected. Borra MT et al, Substrate specificity and kinetic mechanism of the Sir2 family of NAD+-dependent histone/protein deacetylases.(2004) Biochemistry 43: 9877-9887.
(B) Transition State: A novel intermediate-ADPR-peptidyl-imidate intermediate-alpha-1’-O-alkylamidate intermediate has been identified. Asynchronous SN2 mechanism of NAD+ with acetyllysine nucleophile was discussed. X-ray structure data of the transition type analogues on the active site of sirtuins and a QM/MM study of Sir2Tm support this concept. Hawse, WF et al. Structural insights into intermediate steps in the Sir2 deacetylation reaction. Structure (2008) 16: 1368-1377. Hu, P et al. Highly dissociative and concerted mechanism for the nicotinamide cleavage reaction in Sir2Tm enzyme suggested by ab initio QM/MM molecular dynamicssimulations. J Am Chem Soc 2008, 130:16721-16728. A computational study of the Sir2Af2 catalyzed reaction of NAD+ with acetyllysine reached a similar conclusion, fortified with experimental kinetic isotope effects (KIE) data (calculated for candidate transition state structure using computational methods like Gaussian 03 and ISOEFF 98). Cen, Y et al. Transition state of ADP-ribosylation of acetyllysine catalyzed by Sir2AF2 determined by KIE and computational approaches. JACS(2010) 132:12286-12298.
(C) Base Exchange: Using HPLC with [Carbonyl-14C]NAM and 18O-NAM, the base exchange reactions on ySir2, Sir2Af, and mSir2 (SIRT1) were studied. A strategy for increasing Sir2 enzyme catalytic activity in vivoby inhibition of chemical exchange. NAM and isoNAM activation of Sir2 deacetylase activity is achieved without affecting substrate or NAD+ binding by altering the proportion of imidate-enzyme complexes proceeding toward the deacetylated product. A.A. Sauve, V.L. Schramm, Sir2 regulation by nicotinamide results from switching between base exchange and deacetylation chemistry, Biochemistry 42 (2003)9249–9256. A.A. Sauve, R.D. Moir, V.L. Schramm, I.M. Willis, Chemical activation of Sir2- dependent silencing by relief of nicotinamide inhibition, Mol. Cell 17 (2005) 595–601.
(D) Spontaneous non-enzymatic equilibration 2’AADPR_3’AADPR:The mechanism of acetyl transfer to NAD+ includes (1) ADP ribosylation of the peptide acyl oxygen to form a high-energy O-alkyl amidate intermediate, (2) attack of the 2’-OH group on the amidate to form a 1’,2’-acyloxonium species, (3) hydrolysis to 2’-AADPR by the attack of water on the carbonyl carbon, and (4) an SIR2-independent transesterification equilibrating the 2’- and 3’-AADPRs. Approachs used:1H NMR, 2D NMR, MS/MS, 18O exchange, and HPLC. Sauve AA et al. Chemistry of Gene Silencing: The Mechanism of NAD+-Dependent Deacetylation Reactions. Biochemistry (2001) 40: 15456-15463. Smith, BC et al. Sir2 protein deacetylases: Evidence for chemical intermediates and functions of a conserved histidine. Biochemistry (2006) 45: 272-282.__
RC: We will need to indicate which rate constants in the diagram these correspond to. In general, please try to map what you are reading to the models we are developing (not enough to only quote the relevant literature).
XG(8/6): map_reported sirtuin kinetic studies.pptx
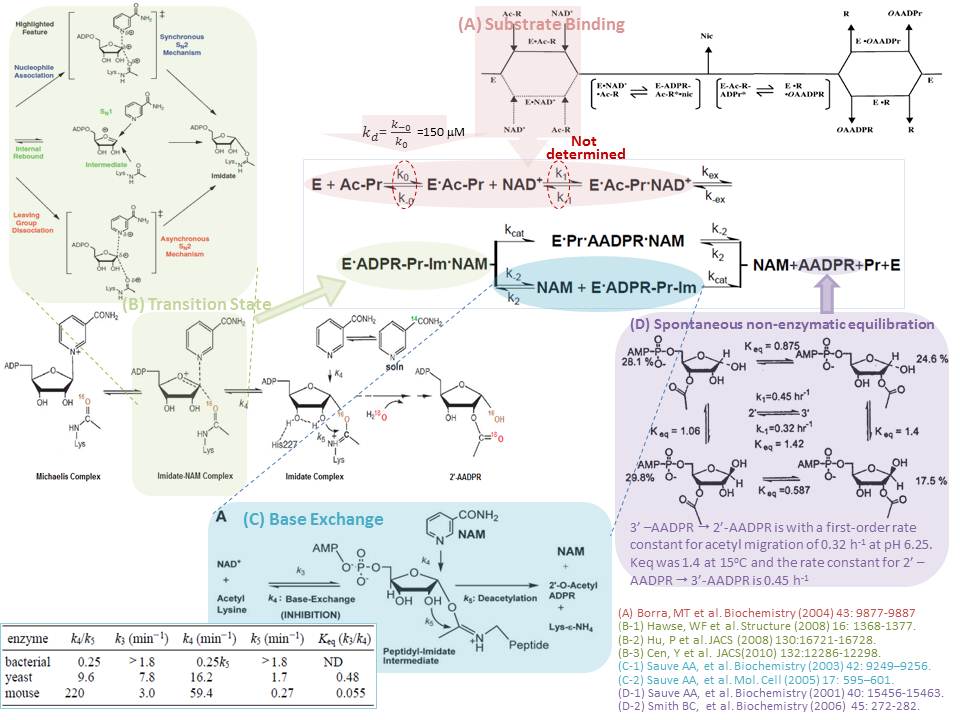
Substrate specificity and kinetic mechanism_2004_Borra.pdf
Structural insights into intermediate steps in the Sir2_2008_Hawse.pdf
Transition state of ADP-ribosylation of acetyllysine catalyzed by Archaeoglobus fulgidus Sir2 determined by kinetic isotope effects and computational approaches_2010_Cen.pdf
Sir2 regulation by nicotinamide results switching between base exchange_2003_Sauve.pdf
Chemical activation of sir2 dependent silencing by relief of nicotinamide inhibition_2005_Sauve.pdf
Sir2 protein deacetylases evidence for chemical intermediates_2006_Smith.pdf
Chemistry of gene silencing_2001_Sauve.pdf
Highly dissociative and concerted mechanism for the nicotinamide cleavage reaction in Sir2Tm enzyme suggested by ab_2008_Hu.pdf
(2) you mentioned “To obtain the rate constant (k), the plot of product concentration formed over time was fitted to a single-exponential equation and then After formation of the ternary complex, nicotinamide is cleaved at a rate of 7.3 s-1 and subsequent formation of OAADPr occurs at 1.3 s-1. “ What is the relationship between these two rate constants and the rate constant k mentioned above?
XG(8/4): The NAM and OAADPr formation rate constants are 7.3 and 1.3 s-1 respectively. They are calculated from the plot using aforementioned method.RC: Not sure we are talking about the same thing here. There was a rate constant k you mentioned. Now you mentioned two rate constants. Do you mean that a single exponential fit is done for the formation of two different products and then two different k's are obtained? Please indicate in the diagram which rate constants we are referring to in each case.
XG(8/6): Two exponential fits are done for the formation of NAM and OAADPR respectively. Two different k’s are obtained. Using a quench-flow apparatus, the HST2 reaction under single-turnover conditions was monitored.
- To determine k_NAM (open square), [14C]NAD+ was used. Fractions from reversed-phase HPLC were subjected to scintillation counting to determine the amount of [14C]nicotinamide formed and [14C]NAD+ left. An average rate of 7.3 s-1 was determined from three separate experiments.
- To determine k_OAADPR (solid circle), [3H]AcH3 was used as a substrate. The amount of OAADPR formed and the amount of nicotinamide formed vs log time were fitted into a single-exponential equation to determine the rate of product formation. An average rate of 1.26 s-1 was yield from two experiments.
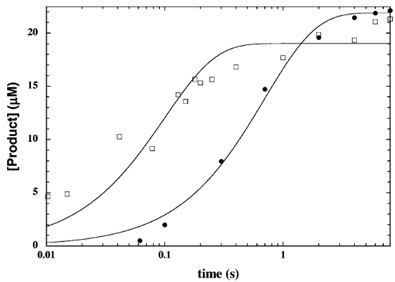
(3) Regarding HPLC, you mentioned at the group meeting that this could be used in post-steady state kinetics (i.e., the transient kinetics that occur after the initial stages of the reaction where steady state conditions no longer hold). Can you provide more details? These discussions will eventually need to be integrated with the relevant kinetic characterization sections of the outline I sent for paper 2.
XG(8/4): HPLC-based Deacetylase Assay relies on the separation of substrates and products of the deacetylase reaction by a reversed-phase HPLC. Quenched reaction mixtures are injected onto a C18 column and, using a gradient of increased levels of organic solvent, substrates, products, and enzyme can be resolved. This assay is applicable to all three classes of deacetylases. For characterization of most Sir2-like enzymes, monoacetylated H3 and H4 peptides, corresponding to the 20 N-terminal residues of histone H3 and H4, can be used. The 110-ul reactions are carried out at 37oC in 50 mM Tris (or phosphate), pH 7.5, with 1 mM DTT. NAD+ and acetylated peptide are mixed and pre-incubated in a 37oC water bath for 5 min. Typical concentrations of NAD+ and acetylated peptide have ranged from 0.25 uM to 1 mM; however, the range should be determined empirically, as the Km values for these substrates may differ by orders of magnitude depending on the Sir2 homologue. The reaction is initiated by the addition of enzyme. The reaction is quenched by the addition of TFA to a final concentration of 1%. Quenched samples are kept on ice or stored in -20oC if not immediately injected onto the HPLC column. Samples are injected onto a reversed-phase HPLC column (e.g., a Vydac C18 column, 4.6 X 250 mm, 201SP104) to resolve substrates and products. A 100-ul loop is typically used for the injections and the flow rate is set to 1 ml/min. After injection, the system is run isocratically with solvent A (0.05% TFA/H2O) for 1 min followed by increasing levels of solvent B (0.02% TFA in acetonitrile). The gradient used for each assay may vary depending on the type of peptide substrate used. For efficient separation of reactions containing the 20-mer AcH3 N-terminal peptides, a gradient of 0–20% B over 20 min is used. For efficient resolution of reactions containing the 20-mer H4 peptides, the following gradient is used: 0–10% solvent B for 4 min followed by 10–25% B for 25 min. Following each run, the column is washed with 100% B for 3–5 column volumes followed by re-equilibration with solvent A for 3–5 column volumes. Elution of substrates and products is monitored by measuring the absorbance at 214 nm (to monitor all substrates and products) or at 260 nm (to specifically monitor nicotinamide, NAD+, and OAADPr). Using the above gradients, a good resolution between the monoacetylated and deacetylated peptides can be achieved. Deacetylated H3 peptide typically elutes at 16 min, acetylated H3 at 18 min, deacetylated H4 at 15 min, and acetylated H4 at 17 min. Substrates and products elute at the approximate percentages of solvent B: nicotinamide at 5%, deacetylated H3 at 16%, deacetylated H4 at 13%, acetylated H3 at 18%, and acetylated H4 at 14%, NADþ at 12%, and OAADPr at 8%. The areas of the peaks are integrated for quantification. To calculate the percent deacetylation, the area of the deacetylated peptide peak is compared to the combined areas of the acetylated and deacetylated peptide peaks. Because a known amount of acetylated peptide is used, the percentage of the deacetylation is then used to determine the amount of deacetylated product formed over the particular time of the assay, to obtain an initial rate.
This HPLC-based assay is also applicable for separating radiolabeled substrates and products. For example, [3H]acetylated histone peptides or [14C]- or [32P]-labeled NADþ can be utilized. [14C]NAD+ can be synthesized using the Sir2-catalyzed nicotinamide-NAD+ exchange procedure (described later) or purchased commerically, and [32P]NAD+ can be obtained from NEN Life Science Products (800 Ci/mmol). The amounts of the radiolabeled substrates and products can be monitored and quantified by collecting the fractions eluted from the HPLC, adding a constant volume of each fraction into scintillation vials and determining the radioactivity of each fraction by scintillation counting. To calculate the amounts of product formed, the radioactivity of the product can be divided by the total radioactivity of all the fractions collected, which corresponds to the radioactivity of the substrate prior to the reaction. The percent product is then multiplied by the concentration of radiolabeled substrate to obtain the concentration of product formed. Alternatively, the total radioactivity of the product can be divided by the specific activity (CPM/mol) of the radiolabeled substrate to obtain the moles of product formed.
One of the limitations of this HPLC-based assay is its relatively timeconsuming nature, where each HPLC run can take approximately 1 h. Only a limited number of injections can be performed over a typical working day. Use of an auto-injector can greatly facilitate the analysis of a large number of samples. Another limitation of this assay is the detector’s inability to detect low concentrations of substrates when absorbance is used to quantitate levels of deacetylation. With low peptide concentrations (below 5 uM) larger injection volumes (e.g., 2 ml) are required in order to have sufficient substrate and products to accurately detect. Moreover, because the retention times may change over column useage, it is necessary to check the elution of standards routinely.
(4) Note that the approach to kinetic characterization in our 2nd paper outline provides a quantitative method for calculating all the rate constants represented in our reaction scheme (which omits certain chemistry and product dissociation steps). Given the posting above, can you consider how to insert bullet points regarding the literature methods in the appropriate sections of the outline that demonstrates how our method offers this advantage? E.g., Sauve's base exchange methods involve various approximations that we do not employ. At least one other slide shows the assumption Kd=Km, which we do not apply.
XG(8/6): 1 day
(5) Please consider bullet points that can be added to the outline regarding the disease applications of inhibitors.
XG(8/6): 1-2 days.
(6) Recall the question regarding the cost of HTS. I was referring to CRO HTS wherein there is a cost quoted per well. This depends on the application, but we can check some approximate prices. Are our assays capable of being run by a CRO in higher throughput, or are they too complicated?
XG(8/4): Few well established companies provide such services. Here two of such companies are listed, Charles River Libraries and Jackson Labs. Here are the services provided by Charles River Libraries:
- Target discovery & validation: Hit identification / Hit-to-lead/Lead optimization / Academic engagement initiative (Grant application support, track record)
- Assay development & screening: Biochemical to Phenotypic assay development/ cell line generation / HTS/High-content screening/ compound screening libraries/orthogonal screening platforms/ion Channel screening/fragment-based drug discovery/novel biomarker identification
- Synthetic & medicinal chemistry: Medicinal chemistry/computer-aided drug design/compound library design/synthetic chemistry/analytical chemistry/process & scale-up chemistry
- Structural biology & biophysics: protein production/protein crystallization &x-ray crystallography/crystal bank/biophysics/fragment-based drug discovery
- Pharmaceutical sciences & formulation: analytics & structural analysis, physical property measurement, solubility, dissolution & stability, salt screening, polymorphism screening, formulation & excipient compatibility.
(7) Regarding the literature on HTS for sirtuins, for the major classes of sirtuin inhibitors/activators currently in pharma development, please list how they were discovered (library type, size), approximate cost based on the prices above, and how they are being optimized (if any info is available, including what types of substitutions are being screened).
(8) Please see the paper in PMC-AT Research dropbox called SRT2104_mouse_lifespan_2014. Can a CRO do preclinical studies of this type?
(9) Indicate when the continuous assay will be ready
XG:
- PCR amplification of PNCA from Samonella genomic DNA (Primer design, PCR condition optimization, …)
- Subcloning into appropriate expression vector-pGEX6P3. (TOPO cloning, transformation of TOP10 chemical competent cell, identification of positive clones, Mini and Midiprep for sequencing, Digestion, Alkaline phosphatase, Calf Intestinal_CIP for Ligation, …)
- Plasmid maxi preparation
- Linearism the constructs
- Transformation the linear construct into the appropriate host strain BL21(DE3)
- Confirmation of positive transformants
- Expression optimization and confirmation. (Screening of transformants, media formulation and inducer concentrations, induction temperature and length, culture lysate conditions, SDS-PAGE gel, …)
- Large-scale expression.
- Purification and confirmation.
- Activity measurements
RC: Time required?
XG(8/4): The experiment was stopped at the purification step. It will take 2 weeks for purification and confirmation and 1 week for activity measurements.
(10)Indicate the approximate amount of time that would be required to get the base exchange assays set up
XG: Most time consuming of the set up process is the purchasing of Liquid scintillation analyzer needed for the experiment. Other than that, we will need a separate room for storage and handling of radioactive reagents. Lab members need to be trained as well (can go on line and check where this kind of training is provided).
(11)Provide a pdf of the final paper with inline figures on the dropbox.
RC: Please prepare a schedule for the above. 11 should be done early next week.
XG(8/4): pdf file named as "PLOS ONE resubmission with figures and tables_08.04.2014" has been created in the dropbox.
Dropbox/PMC-AT Manuscript/PCB-SIRT3 inhibition/PLOS One resubmission/
XG(6/13/14): PMC-At group meeting follow up_part one is attached. Will continue working on the completion of all the tasks.
PMC-AT group meeting follow up-part I.pptx
RC: Comments:
i) you mentioned that
Previous studies have focused on the complete steady-state kinetic mechanism
üthe inherent differences in catalytic efficiency and substrate preference among Sir2 enzymes (ySir2, yHST2, and SIRT2) for various histone peptides
üthe overall kinetic mechanismüResolve the individual chemical steps of the Sir2 reaction.Specifically in terms of the rate constants of the reaction mechanism, what does mean? In what way were each of the individual chemical steps resolved? Subsequently, you mention that rapid quench was required to determine particular reaction rates.
ii) you mentioned
To obtain the rate constant (k), the plot of product concentration formed over time was fitted to a single-exponential equation and then After formation of the ternary complex, nicotinamide is cleaved at a rate of 7.3 s-1 and subsequent formation of OAADPr occurs at 1.3 s-1. What is the relationship between these two rate constants and the rate constant k mentioned above?
iii) Regarding HPLC, you mentioned at the group meeting that this could be used in post-steady state kinetics (i.e., the transient kinetics that occur after the initial stages of the reaction where steady state conditions no longer hold). Can you provide more details?
These discussions will eventually need to be integrated with the relevant kinetic characterization sections of the outline I sent for paper 2.
XG(6/20/14):In terms of the rate constants of the reaction mechanism, some of them for individual step(s) have been measured experimentally or computationally. The methods are listed in the following file. Sirtuin chemical mechanisms_06.20.2014.pptx
RC (6-25):
a) Note that the approach to kinetic characterization in our 2nd paper outline provides a quantitative method for calculating all the rate constants represented in our reaction scheme (which omits certain chemistry and product dissociation steps). Given the posting above, can you consider how to insert bullet points regarding the literature methods in the appropriate sections of the outline that demonstrates how our method offers this advantage? E.g., Sauve's base exchange methods involve various approximations that we do not employ. At least one other slide shows the assumption Kd=Km, which we do not apply.
XG(6-25): Will do.
b) As noted please examine the list of leads Ping has identified so far (e.g. the highest ranking mimics, using the ranking at higher level of theory) and check whether they violate the patents on Ex-527. If not, please start running them through the similarity search in the commercial databases to see what are the closest molecules we can buy. Then Ping can rerank those to check the change in rank for commercially available molecules.
Then please comment on how these leads would be tested in the lab, including how many could be tested in a month.
XG(6-25):Ping currently has the selection of leads (70,000) targeting three different receptors (product, intermediate, and SIRT3:NAD+). Most of the work has been done on product and SIRT3:NAD+. First step, the top 200 leads targeting product will be picked up. The general procedure is listed below:
*ChemBridge Molecule Store and eMolecules
*draw the molecular structure
*run similarity search
*pick the candidates from output structures
Basically it will take 10-15 min per lead. Please advise if 200 molecules are too much.
RC: That sounds ok. Please provide me with some of the structures with comparison to the computational leads asap.
RC (8-4): Please provide me with an update and list of next steps for the similarity search and validation of the leads (including crosschecking the leads against the patented structures) today, so we can settle the schedule for this work. Please include a status update from PL on the rescoring calculations for this.
XG(8/4): similarity search_8.4.14.pptxresults from similarity search_8.4.14.docxThe MMGBSA dG binding of the top 20 molecules are listed. The next steps are including
(1) patent search (not sure how long it will take).
(2) compound ordering
(3) The first round of similarity search is based on the MOE CoreHopping results with aADPR as receptor (mimicking Ex-243). See other possibilities, different receptor? different mechanism
RC: Please start with the patent search on the paper I gave you yesterday on pyridine derivatives - please complete this today. Also today, please check whether it is mentioned anywhere in that paper what the NAM concentration was in the in vitro assays (check supporting info as well).
XG(8/5): The compounds mentioned in J Med Chem-2009 paper have been included in patent US2007/0161683 A1. Compound 1c is commercially available.
J Med Chem_2009 compound patent search.pptx
XG(8/5): No additional NAM was added in the assay. The absolute value of SIRT1/2/3 activity was measured. The follow the equation (below) to calculate % SIRT activity.
Fo=Absolute value of SIRT1/2/3 control
Fn=Absolute value of SIRT1/2/3 activity with modulator
% SIRT activity= 100 *Fn/Fo
Fluor de Lys fluorescent biochemical assay was applied and no kinetic study was involved.
2009 J Med Chem Qustion.pptx
c) Please consider bullet points that can be added to the outline regarding the disease applications of inhibitors
d) A couple of questions above (e.g. ii) seem to still be pending
XG(6-25): Will do.
Lower priority:
e) Recall the question regarding the cost of HTS. I was referring to CRO HTS wherein there is a cost quoted per well. This depends on the application, but we can check some approximate prices. Are our assays capable of being run by a CRO in higher throughput, or are they too complicated? Also, regarding the literature on HTS for sirtuins, for the major classes of sirtuin inhibitors/activators currently in pharma development, please list how they were discovered (library type, size), approximate cost based on the prices above, and how they are being optimized (if any info is available, including what types of substitutions are being screened).
f) Please see the paper in PMC-AT Research dropbox called SRT2104_mouse_lifespan_2014. Can a CRO do preclinical studies of this type?
XG(6-25): Will do.
RC (6/3/14): Please post the minutes for your next tasks from the group meeting here, including action items proposed by all attendees.
These include:
a) further comments on transient kinetics - please include a summary of burst phase. Also, use of HPLC analysis in this context.
b) looking up pathways whereby sirtuin inhibition can alleviate metabolic or neurodegenerative disease
c) listing the increase in throughput due to use of continuous assay and parallel experiments
d) proposed plan
for continuous assay characterization steps
e) cost estimate of experimental HTS from CROs
a,c) may be useful for writing the proposed experimental methods section for the 2nd paper. Base exchange experiments will also be discussed there.
If we can use a similarity search to identify molecules from a commercially available library similar to the computational hits, these may be used
to develop a training set for computational binding affinity predictions, and hence new experiments with such a congeneric series of extended C pocket ligands
will be done. You can help with the similarity search.
RC (6/9): Here are some articles on the disease applications of sirtuin inhibitors; most refer to SIRT1. Neurodegenerative diseases appear to be a prime target, with Ex-527
apparently in trials for HD. We need to review this (note preclinical animal models used by Elixir for HD) and summarize main points, and check whether anyone is targeting SIRT3, given that it is a "double-edged sword".
http://www.physicventures.com/news/examining-both-sides-sirtuin-coin examining both sides of the sirtuin coin_2010_Dimond.pdf
http://www.sciencedirect.com/science/article/pii/S0925443908000616 Therapeutic role of sirtuins in neurodegenerative disease_2008_Outeiro.pdf
http://www.nature.com/nrd/journal/v7/n10/execsumm/nrd2681.html Therapeutic application of histone deacetylase inhibitors for central nervous system disorders_2009_Kazantsev.pdf
http://www.ncbi.nlm.nih.gov/pubmed/20872319
XG(6/9): OK.
XG(5/27/14):
PMC-At group meeting Outline
RC (5-27): Regarding Tecan, please indicate whether/how it can be used for transient kinetics experiments in addition to steady state experiments (this pertains
to our interest in mechanism).
Please have one backup slide with some of the results from N-methyl NAM experiments.
Please post the draft ppt sections as they become available (not necessarily final versions).
XG(5/28/14): OK.
XG(5/7/14)
(1) IC50 of Ex527 for SIRT3 and SIRT1 have tested (4/24/14-5/2/14).
(2) Ex527-SIRT3 kinetics – 25 uM (5/8, 5/9, 5/12/14)
(3) Ex527-SIRT3 kinetics – 50 uM (5/13, 5/15, 5/16/14)
(4) Ex527-SIRT3 kinetics – 75 uM (5/19/14 – 5/21/14)
RC: Please prepare a powerpoint on Ex-527 experiments as well as a review of the experimental results from the PNAS paper. Are you doing any more experiments next week?
Recall there were some discussions regarding the possible use of Tecan to increase the throughput of our lead discovery screening experiments as well as to do transient analysis
of sirtuin kinetics. Please include some info on how the instrument could be used to do these types of experiments. Please indicate whether any additional apparatus would be required
to do pre-steady state kinetic analysis.
Also, I believe you have done some patent analysis. Some of that could be included as time permits, although the emphasis in lit review would be the PNAS paper (esp how they drew inferences
about the mechanism from the experimental data and how they related their binding affinity data to the binding affinity of particular complexes).
You may also include a slide on experimental high throughput screening of the type that was used to discover Ex-527 (PL has the paper) and how costly it is (optional, if time permits).
Please try to send an outline of your ppt within the next day or so, so I can provide feedback if needed and we can determine whether to have the group meeting on Fri or Mon of next week.
XG(5/27/14): Ex527-SIRT3 kinetics (25, 50, and 75 uM) has been completed. I will analyze data on 5/27 and 5/28. Then prepare powerpoint as requested. No more experiments are scheduled this week.
XG(3/31/14)
Schedule for week of 3/31/14-4/4/14
(1) isoNAM-SIRT1 relative inhibition in the presence of 200 uM of NAM
(2) isoNAM-SIRT3 relative inhibiton in the presence of 200 and 400uM of NAM
(3) Test on 1-methyl-NAM Chloride solubility and physio-chemical properties of stock solution
(4) 1-methyl-NAM Chloride-SIRT1 relative inhibition (how many concentration and how high it can go will depend on the solubility test);
(5) 1-methyl-NAM Chloride-SIRT3 relative inhibition (how many concentration and how high it can go will depend on the solubility test).
Dr Raj, experiments (1) and (2) are the extended tests from last week's results. Please advise if they are necessary to do at this point. If not, the experiments will be move to (3).
RC: Yes, we should do relative inhibition studies under higher concentrations of NAM to study derepression of NAM inhibition under conditions of greater inhibition by NAM. The above tasks are approved.
Please make sure to report the results of (1) as soon as there are in and prior to starting on (2). I will provide more commentary on such studies thereafter.
XG(3/24/14)
Schedule for week of 3/24/14-3/28/14
--isoNAM-SIRT1 Relative inhibition (one concentration) (3/24-3/27)
RC: Relative inhibition studies should be continued as discussed. Regarding the SIRT3 relative inhibition experiments, above 1 mM did you use the old or new kit? You did those experiments before the SIRT1 experiments, so I assume you used the old kit. After finishing SIRT1, are you planning to redo any of the SIRT3 isoNAM concentrations as well?
Please provide an updated schedule for these experiments, after which we will look at methylnicotinamide.
XG(3/26/14): Yes. I did isoNAM-SIRT3 relative inhibition and kinetics experiments before the SIRT1 studies. The SIRT3 kits used for those studies were newly purchased and arrived at PMC-At lab on 1/9/14. SIRT3-isoNAM experiments do not need to redo. isoNAM-SIRT1 relative inhibition experiments will be completed on Thursday with data analysis. We have 1-methylnicotinamide chloride in the lab. Start from Friday, the physio-chemical properties (solubility, reaction pH, in assay buffer...) of this chemicals can be tested. Which enzyme (SIRT1/SIRT3) are you interested? I need to prepare the protocol and calculate if we need order more kit.
--continue on literature search during assay preparation and sample incubation (3/25-3/28)
RC: After you finish the current experiments you should repeat a couple of data points with a separate kit - I believe you have one extra new SIRT1 kit. Then for N-methyl-NAM you should prepare to do experiments with both SIRT1 and SIRT3 (SIRT1 first).
XG(3/26): The order of the experiments:
(1) isoNAM-SIRT1 relative inhibition (2 concentrations: 10mM and 100 mM) using new kit;
(2) 1-methyl-NAM Chloride-SIRT1 relative inhibition (how many concentration and how high it can go will depend on the solubility test);
(3) 1-methyl-NAM Chloride-SIRT3 relative inhibition (how many concentration and how high it can go will depend on the solubility test).
XG(3/17/14)
Schedule for week of 3/17/14-3/21/14
--- Kinetics experiment of isoNAM: SIRT1_one concentration (3/21)
--- Literature search
(1) Value of alpha if provided for mixed inhibition fitting to all sirtuins studied previously
(2) Prepare a detailed ppt on the state-of-the-art knowledge regarding NAD+ and NAM concentrations in celluar subcompartments.
(3) a patent and literature search on NAM-derepressant small molecules in addition to isoNAM
(4) From Dr Raj: NAM has been reported to have numerous health benefits: A paper in Nat. Chem. Biol 2013 showed NAM can increase lifespan of worms. More general health survey of NAM: http://www.resveratrolnews.com/history-of-nicotinamide-and-aging/868/
(5) From Dr Raj: effects of NAD+ on health (related to task on NAD+/NAM levels in cell): One article recently read indicates that PARP and CD38 inhibition can help boost NAD+ levels:
"It is of note that the NAD+ boosting effects of PARP inhibition enhance the activity of SIRT1, but not that of SIRT2 or SIRT3 (Bai et al., 2011b). The major difference between these three sirtuins is their subcellular localization, because, among them, only SIRT1 is a nuclear sirtuin. This suggests the existence of compartment-specific NAD+ pools in the cell. Supporting this possibility, elegant studies by Yang et al. (2007a) showed the existence of independently regulated NAD+ pools. CD38...."
RC: Please provide expected completion dates/schedule for the tasks.
XG(3/21): the literature review will take longer time than what I expected. The aforementioned 5 topics have cross-talk and a lot of reading is needed. Currently, I have finished part of (2) and (3). Next week the focus will be on isoNAM-SIRT1 kinetics experiments. I will use the time between experimental setting-up and sample inculation to do more search.
RC: I mean including the experimental tasks. Were any experiments carried out this week? Please provide the detailed schedule going forward.
XG(3/21): IsoNAM-SIRT1 kinetices one concentration will be finished today, and next week(3/24-3/28) will do another 2 concentrations. Then 2 more days data analysis and model fitting
RC: Do you mean relative inhibition? I thought you were planning to repeat one concentration of isoNAM:NAM:SIRT1 relative inhibition.
XG(3/21): IsoNAM-SIRT1 relative inhibitionis planning after the kinetics finished. If the relative experiments need to be done first, then I can do it first next week.
RC: I believe we discussed that they should be done first. This is because if the observed activation does not occur, the priority of experiments may change.
Ideally the single concentration of isoNAM would be the one that produced maximum activation in the original dataset. When do you plan to investigate the issues observed with higher [isoNAM] to resolve if that was a real effect of experimental artifact?
XG (3/24): To resolve the experimental artifact issue, another assay method needs to be applied. The comparison of strengths and weaknesses of the well-established Sirtuin activity assays is listed as following:
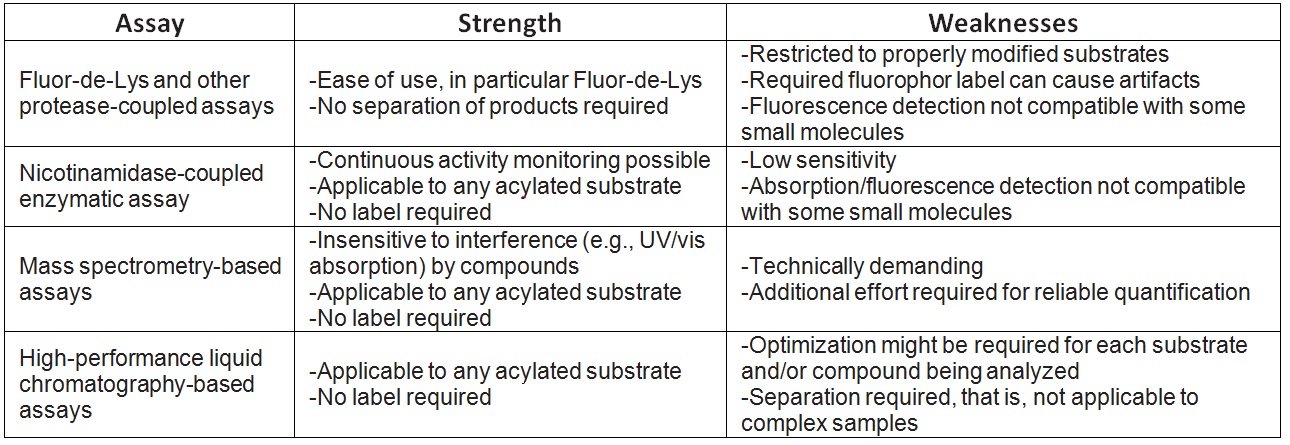
For the current availabilities of the instruments in our lab, HPLC based assays might be the reasonable starting point. Depending on the priority of the pending tasks (isoNAM-SIRT1 kinetics, PNC1-coupled enzymatic assay, literature search), I need further advice on when we can investigate this issue.
CJ (3/10/14)
Schedule for this week:
3/10/14 - 3/11/14 Try a fusion PCR to assemble f1 - f3 to construct the P99 library. This is just a proof-of-principle experiment to make sure the fusion PCR protocol works. The assembly of the real library will be done after the Tecan arrives, so that the DNA concentration can be measured accurately. The Tecan is expected to arrive next week.
3/12/14 and 3/14/14 Review Karthik's report and figure out how to improve experiments for the next step. During our last meeting Karthik promised to send me some simulation results by last Thursday, but I'm still waiting for them. Meanwhile I will also communicate with Chun-Li Chang on the COLD-PCR collaboration.
3/13/14 The CCG workshop.
XG(3/3/14)
Schedule for week of 3/3/2014-3/7/2014
v Tasks related to manuscript revision
--As previously noted, revised figures with the error bars shown.
--A table reviewing the literature data on the Km's of various sirtuin (for discussion)
--Methods para for the global nonlinear fitting
--Look up NAD+ physiological concentrations - including in cellular subcompartments, if available (mitochondria of interest, but unlikely to be known). Any available data on NAM as well.
--Look up IC50 of NAM for SIRT5.
-- updating the citations in the manuscript
v Experiments plan
--Compare the maximum concentration of [NAM] used to that used by Sauve 2003 in the Dixon plots.
--Estimate how long it will take to run a series at constant saturating [NAD+] for various [NAM] concentrations for Dixon plot.
--Redo one [isoNAM] for SIRT1
RC: The isoNAM task is the highest priority among experiments. Please try to do it some time this week.
CJ (2/28/14)
I missed one time point during the extension assay on 2/25/14 while talking to Sherry about the email issue. I will repeat part of the experiment next week.
My schedule next week:
3/3/14 agarose gel electrophoresis of f3 for beta-lactamase library. If bands are good, will then purify DNA and send for sequencing.
3/4/14 - 3/5/13 repeat part of extension experiments for fixing [N] at 72C, [SP0]=2 - 60nM.
3/6/14 - 3/7/14 on vacation. (Sherry has signed my PTO form.)
CJ (2/24/14)
My schedule this week:
2/24 - 2/25 extension experiments for fixing [N] at 72C, [SP0]=2 - 60nM
2/26 - 2/27 extension experiments for fixing [N] at 72C, [SP0]=80 - 200nM
2/28 fluorescence measurements and data processing
RC (2/23/14): a) Please run 1-2 higher concentrations (above 100 mM) of isoNAM in the SIRT1 relative inhibition experiments.
XG(2/24/14): 100 mM isoNAM reaction solution need to be prepared from 1 M isoNAM working solution which is a bit concentrated. To go even higher, the DMSO may be introduced for better solubility. Will try and may take 3 days.
b) Please comment again on the issues with running additional concentrations of NAM/NAD in the NAM inhibition kinetics experiments for SIRT1. If there is an issue with comparability of data, we will not do this.
XG(2/24/14): I can run one set of experiment in which all the concentrations are included. Then use standard curve to calculate and find the constant. For the rest of NAM concentrations, the experiment will be performed only at the additional NAD concentration. Then convert the data into original curve. It will take 5 days. Please comments (1)how many additional concentrations of NAD we need to run? (2) what the concentration range is?
RC: It will be best if you can first provide the information regarding the enzyme concentrations mentioned earlier and also comment on whether there were certain concentration ranges (e.g. high NAM, low NAD) that were skipped in the SIRT1 experiments but done in the SIRT3 experiments. I recall you did one fewer NAD and NAM concentration for SIRT1 vs SIRT3. Also, please comment on whether you think there may be problems comparing the old and new data. If so, we may not do these experiments now. If you feel the omitted NAD concentration and one NAM concentration were in the low NAD, high NAM range, and the data will be comparable, let me know.
In any case, you can do a) above first while you assess this. However, we will most likely not use a) in this paper.
c) Plan to start running the isoNAM/SIRT1 inhibition kinetics experiments. These will not be used in the current paper, but will be used in the next one.
XG(2/24/14): Will plan. For 3 isoNAM concentration and 5 NAD concentration, it will take 12 days for experiments and 2 days for data anaysis.
d) While doing c) please finalize your figures and write up your methods.
XG (2/24/14): OK.
RC: Since you may not be starting c this week (given that a will take 3 days), please make sure you work on d) this week.
XG (02/17/14)
Schedule for week of 2/17/14-2/21/14
2/17/14 Data analysis (isoNAM-SIRT3 kinetics experiment)
Dr Raj, please prioritize the following tasks
-- IsoNAM-SIRT1 kinetics experiment (12 days for experiments and 2 days for data analysis)
-- IsoNAM-SIRT1 relative inhibition experiment (3-4 days)
RC: Relative inhibition of isoNAM-SIRT1 should be done first. Once I receive the relative inhibition results
for isoNAM-SIRT1, I will comment on next steps. You might consider doing higher isoNAM concentrations first (around the IC50, if you know it) and sharing that data so we can check activation, since that may affect paper writing.
Please let me know how many NAD concentrations the above kinetics experiment includes. Is it possible to sample low/high concentrations first at the three isoNAM concentrations and then do more? In particular, are you planning to do the same number of concentrations for isoNAM-SIRT1 as previously done for NAM/SIRT1? It would be odd to do more concentrations for isoNAM-SIRT1 than you did for NAM-SIRT1.
XG(2/17): The reason we measured less NAD concentrations (4 [NAD]'s) on NAM-SIRT1 kinetics experiments was because it's well known. The purpose we included NAM-SIRT1 data was to show our method can succefully repeat published results. On the case of isoNAM-SIRT1 experiments, this is a novel study and I prefer to do 5 NAD concentrations for the determination of initial reaction rate.
RC: So you mean 3 isoNAM concentrations and 5 NAD concentrations?
XG(2/17): Yes.
RC: Please provide the following additional information: a) what is the concentration of enzyme used in these experiments, and how does the range of [NAD]_0/[E]_0 ratios used in these experiments compare to those used in the literature on Sir2.
XG (02/18/14)
-- For SIRT3 experiments, 5 NAD concentrations (100, 375, 750, 1500, and 3000uM) were applied on all the experiments. [SIRT3] = 2.5 U (1U=1 pmol/min at 37oC with 500 uM substrate peptide and 500 uM NAD).
In Sinclair-Sirtris 2008 paper (Biochemical characterization, localization, and tissue distribution of the longer form of mouse SIRT3), 1.47 nM mSIRT3L-54-334 was used with a fixed substrate peptide concentration of 2 uM and [NAD]=16, 31, 63, 125, 250, 500, 1000, 2000 uM.
-- For SIRT1 experiments, 4 NAD concentrations (50, 125, 750, 1500 uM) were used. [SIRT1]= 1 U (1U = 1pmol/min at 37oC with 250 uM substrate peptide and 500 uM NAD).
In Sauve 2003 Biochemistry paper(Sir2 regulation by NAM results from switching between base exchange and deacetylation chemistry), 1 uM Sir2 enzyme was used with 300uM substrate peptide and 600 uM NAD containing selected micromolar concentrations of [carbonyl-14C]NAM at 60 uCi/umol (0, 10, 20, 30, 45, 60, 80, 90, 125, 250, 360, 600, and 1200).
I noticed that the lowest [NAD] you used was 1/4 of the next, and in the other cases you doubled the [NAD] for each successive point. I am asking because I would like to verify that these are the concentrations we should use in future experiments as well.
b) How many experiments is each reported data point based on.
XG(02/18/14): Single experiment, no repeat.
(Please bear in mind that whenever possible in the future, after each series of data is collected in such experiments. I would like you to carry out and report data analysis so it is easier for me to advise in real time how to allocate time to additional concentrations or new experiments.)
-- One additional low [NAD] data point for SIRT1 inhibition by NAM at all [NAM]'s (5 days)
-- Write up the modified Prism fitting methods (for methods section) based on the new global fitting method
CJ (2/17/14)
My schedule this week:
2/17 - 2/18 extension experiments for fixing [SP0] at 72C, [N]=300 - 1000uM. (I was not able to finish this experiment due to the office closing last week.)
2/19 fluorescence measurements and data processing
2/20 - 2/21 PCR to amplify f3 for the beta-lactamase library, followed by gel purification and sequencing sample preparation.
CJ (2/10/14)
My schedule this week:
2/10 - 2/11 extension experiments for fixing [SP0] at 72C, [N]=2 - 220uM
2/12 - 2/13 extension experiments for fixing [SP0] at 72C, [N]=300 - 1000uM
2/14 fluorescence measurements and data processing
XG(2/10/14) Schedule for week of 2/10/14-2/14/14
2/10/14-2/12/14 isoNAM kinetics (the third concentration)
RC: Ok, please post the plots from the 1st two concentrations when you get a chance.
2/13/14-2/14/14 Data analysis (Prism inhibition model fitting)
CJ (2/4/14)
My schedule this week:
2/3/2014 analyze sequencing results of the second round of SDM - done
2/4/2014 - 2/5/2014 run PCR and gel purification for fragment 1 (f1) and send for sequencing.
2/6/2014 - 2/7/2014 (1) run PCR and gel purification for fragment 2 (f2) and send for sequencing. (2) analyze sequencing results of f1.
Please see the attachment on 1/23/14 on beta-lactamase wiki for definition of fragment 1 and 2.
XG(2/6/14):
2/6/14: VP-ITC testing and evaluation.
2/7/14: If ITC testing is finished, isoNAM kinetics work will be performed.
RC (2/3/14): XG and CJ should use this page to post their weekly schedule at the start of each week. (That means including this week if you are in the lab.)
Friday_100413
The current projects are (A) SIRT3 inhibitors/activators screening; (B) study of inhibition modes for certain inhibitors. Project A will provide leads for the study of project B, meanwhile, the results of project B will help the understanding and design of drug screening protocol.The bigger picture of our projects has been represented in the Figure 1, which indicates the role of computation in the drug screening and how experimental part evaluates the hits from simulation.
Briefly, a large library of commercially available chemicals is needed for virtual screening. Ping has downloaded a free database which contains over 21 million purchasable compounds in ready-to-dock 3D formats. High Throughput Screening (HTS) will be applied at the first stage and pharmcophore for enrichment by using Glide/Phase 3.6.
When the list is narrowed down, Glide SP, Glide XP, and MM-GBSA calculation will be used on docking step. The successful docked compound will be selected and pass to next round of MD simulation with MM-GBSA possible LRM for more accurate binding affinity calculation.
The drug candidates will be evaluated through experimental methods. (1) Measurement of IC 50 will provide a general idea of the potency of inhibitor. (2) Study of inhibition mode will help to understandthe mechanism. (3) Measurement of binding affinity is necessary and provides solid support for the comparison to simulation work. TLC can be used as one of the methods and the purchase of such an instrument is suggested.
From literature search, we noticed that large portion of the inhibitors follow different type of inhibition (see Table below). These results indicate that IC50 does not always reflect the inhibition binding affinity and in the case it does, inhibition mechanism also affects how we estimate inhibition constant from IC50.
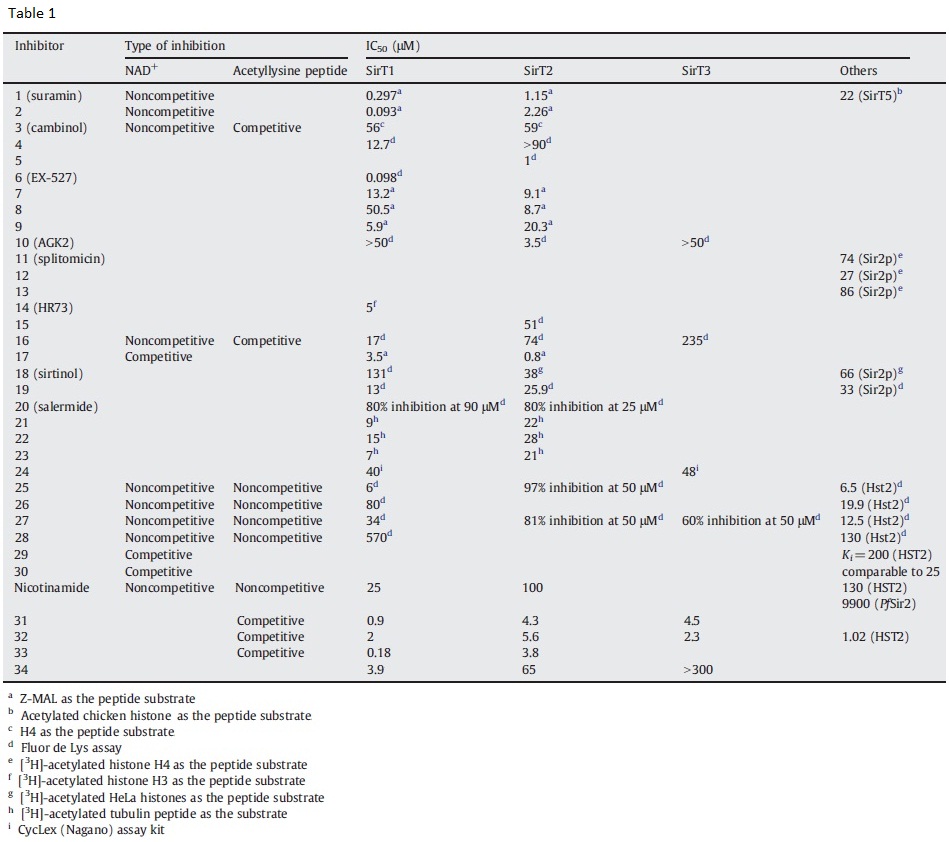
For example, Ex527 is not working by binding to active site but intermediate. Therefore, as shown in Figure1, the Green portion represents the strategies of preparation of receptor according to different inhibition mode: (Type 1) if the drug is targeting NAD+, then SIRT3 with LysAC peptide is used as receptor; (Type 2) if the LysAC is targeted, the receptor will be SIRT3 with NAD+; (Type 3) if the drug is targeting both of NAD+ and LysAC, then Apo-enzyme is used as receptor. The above three is for inhibitor screening. (Type 4) If the drug inhibits the NAM-exchange reaction, the receptor will be SIRT3 with intermediate. Type 4 is specific for activator design.
The plan for aforementioned bigger picture-
Currently we have 12 compounds (NAM, isoNAM, 1-methylnicotinamide chloride, nicotinic acid, pyridine N-oxide, Ex527, salermide, AC93253, CamBridge 5281087, 4102009, 9147724) in the lab, 9 of them (NAM, isoNAM, 1-methylnicotinamide chloride, nicotinic acid, pyridine N-oxide, Ex527, salermide, AC93253) have been tested for IC50 and NAM was tested for inhibition mode. Experimental data show that 1-methylnicotinamide chloride, nicotinic acid, pyridine N-oxide are weak SIRT3 inhibitors, which are small molecules, and can be considered as possible activators like isoNAM through inhibition of base exchange.
Simulation part:
(A) Inhibitor screening
- Current available inhibitors: Ex527, salermide, AC93253, CamBridge 5281087, 4102009, 9147724
- HTS from ZINC
- MD method setup and development
- Current available molecules: isoNAM, 1-methylnicotinamide chloride, nicotinic acid, pyridine N-oxide
- Others
70% of Enzyme-coupled continuous assay has been finished. Need to keep working on it to complete. Because some inhibitors do not really simply follow one specific inhibition mode (competitive, noncompetitive, uncompetitive) as shown in Table 1, it’s important to quench deacetylation reaction in the presence of modulators and capture some intermediate states for mechanism study as well providing enriched info for simulation work.
(A) Inhibitor screening
- Current available inhibitors for IC50 measurement: CamBridge 5281087, 4102009, 9147724
- Study of Inhibition mode: Ex527, salermide, AC93253, CamBridge 5281087, 4102009, 9147724, any others from Ping’s results
- TLC for measurement of accurate binding energy
- Measurement of rate constant of production of intermediate or other interesting products or reactants.
- Experimental methods development for study of Base-Exchange reaction, including NAM on/off rate constant…
- % of Inhibition of deacetylation reaction vs. Concentration of small molecule in the presence of NAM.
Friday_092713
Large-scale expression. Scale up of culture from 10ml to 100 ml. IPTG induced overnight at 37oC. The cultures were harvested and aliquot as different portion for (a) glycerol stock, (b) miniprep, and (c) lysation. All the samples are stored at -80oC for further operations.- Miniprep plasmid will be digested overnight at 37oC (9/30/2013), the digested products will be run on Agarous gel to check for correct target size. By then will decide if sequencing services is needed.
- Sample (c) will be lysated and SDS PAGE will be followed up to check the level of expression and correct size for target protein.
RC: Are you planning to start doing any inhibition mode studies on the small molecules? Please specify the schedule/plan for this vis-a-vis the work of Ping.
XG(10-2):
For inhibitor screening (IC50 measurements to show the potency of inhibition, Ki then can be calculated), it takes 1 month for testing 5 inhibitors using Fluor-de-Lys SIRT3 drug discovery kit (2 kits, ~$1200).
For inhibition mode study (determination of competitive, noncompetitive, uncompetitive inhibition modes), it takes 4 weeks for testing 1 inhibitor using Fluor-de Lys SIRT3 drug discovery kit (3 kits, ~$1800). General schedule are listed below:
Week 1: Inhibitor physiochemical properties
(a) Solubility in H2O, assay buffer
(b) If not soluble or have very poor solubility in assay buffer, selection of organic solvent
(c) Concentration range, pH range
(d) If has color, background minimization
Week 2: Standardization of working assay systems
- Standard calibration curve
- Titration of developer signal stabilization
- Measurement of Km(NAD+) and Km(substrate peptide)
- IC50 (may try 10 different inhibitor concentrations, and then narrow down for specific range.)
- Ki ( [NAD+]=0, 62.5, 125, 250, 500, 1000, 1500, 3000 uM with different inhibitor concentrations.
Ping and I have a short conversation to discuss which inhibitor should be tested. It’s a good starting point to test the inhibition modes of Ex-527, Salermide, and AC93253 for SIRT3. Known that the inhibition modes (noncompetitive, competitive and uncompetitive) are based on simplified model, which may not reflect the complicated case. In another word, up to date, some reported inhibitors do not follow simply one inhibition mode. For example, Ex-527 has been tested on SIRT3 experimentally (crystallography) and computationally (PNAS 2013). It’s reported that Ex527 inhibit SIRT3 followed by another mechanism. Ex527 occupies that NAM site and a neighboring pocket, and stabilizes the closed enzyme conformation preventing product release. It’s been reported that Salermide has a strong in vitro inhibitory effect on SIRT1 and SIRT2. Docking studies show that Salermide can be docked onto the C-pocket of human SIRT2 after minimization (Oncogene 2009).
Therefore, the current experimental methods will provide the inhibition modes for aforementioned candidates. However, if we want to study the inhibition mechanism, we would like to have more experimental data to propose one or few possible modes for Ping to compute. The measurement of quantitation of intermediate/ product will be very useful. The binding affinity measurement by TLC is helpful as well.
Notice that the study of inhibition mode and the development of continuous assay cannot be performed parallel. Please advise the priority of these two assays. I am working on the continuous assay now, will switch if it is needed.
Friday_083013
The IPTG induced method was modified and fixed at the conditions, in which the expression level of pGEX-6P3-PNCA protein is very high and consistent. The efficient cell lysate protocol was achieved to get most of the target protein into soluble portion.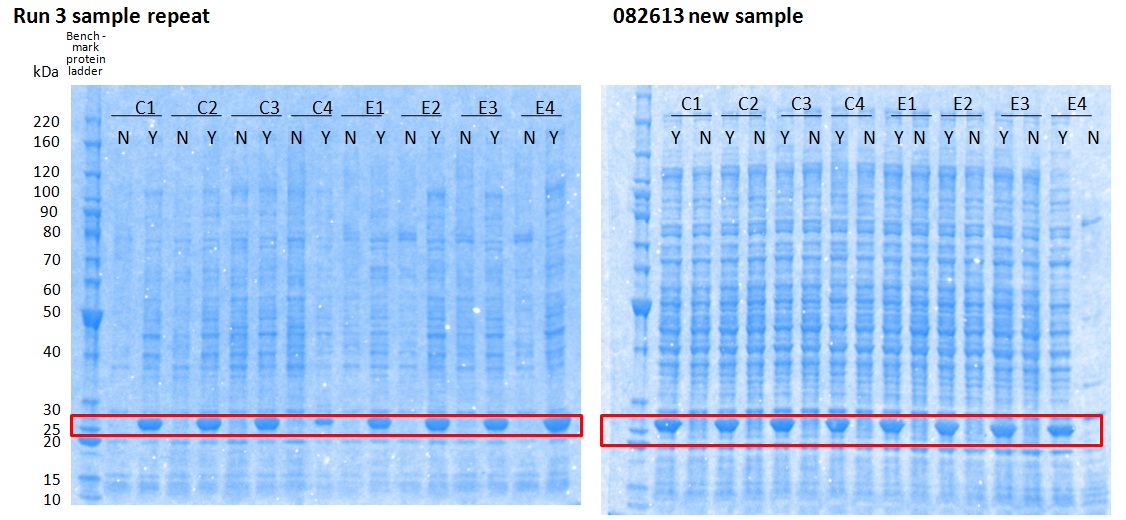
Next step is to scale up the culture amount, large scale protein purification.
Friday_080913
Experiments
Current focus is to obtain high expression level of pGEX-6P3-PNCA protein for continuous assay. In order to reach the goal, different induced condition (titration the concentration of IPTG, induced temperature, and induced time) and cell lysation method (lysate buffers, reagents, sonication details) were optimized.Run 1: The target protein was induced by IPTG and pGEX-6P3-PNCA protein expression was detected (sample E2, E4, E5, E6, C1, C2, C3). However, the expression level was low.
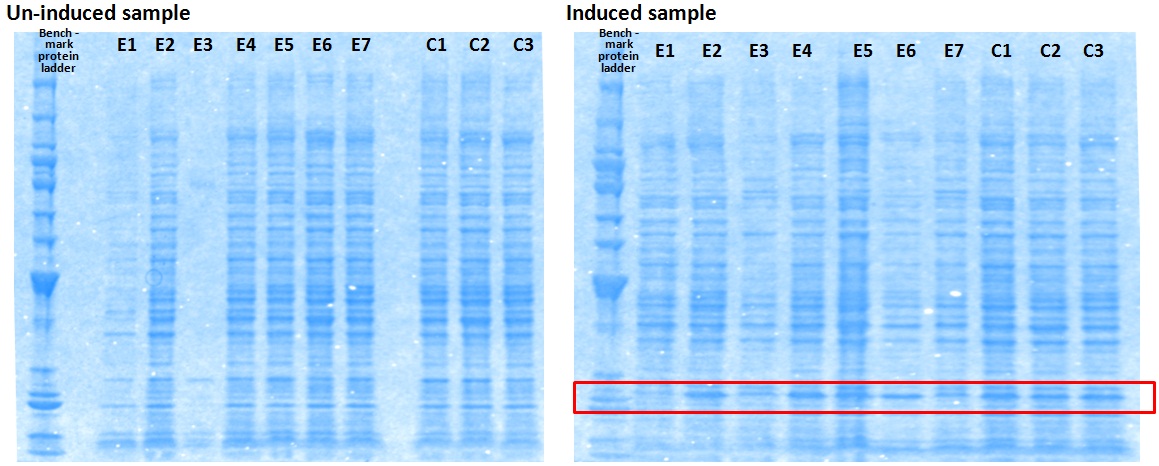
Run 2: The target protein was induced by IPTG and pGEX-6P3-PNCA protein expression was detected (sample E1, E2, E4, E5, E6, C1, C2, C3). The expression level was improved.
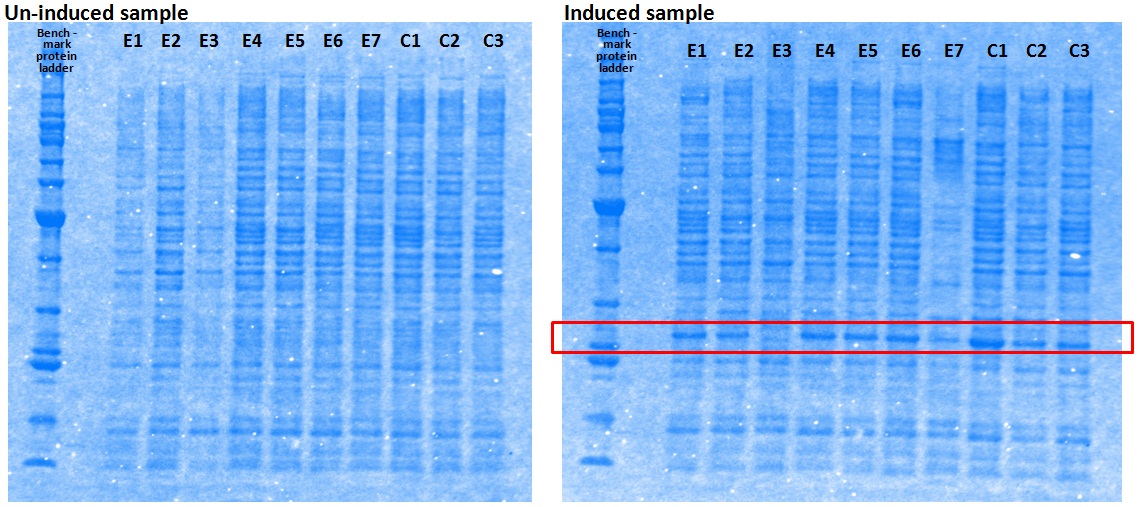
Run 3: The target protein was induced by IPTG and fair amount of pGEX-6P3-PNCA protein expression was obtained (sample E1,E2, E3, E4, C1, C2, C3, C4).
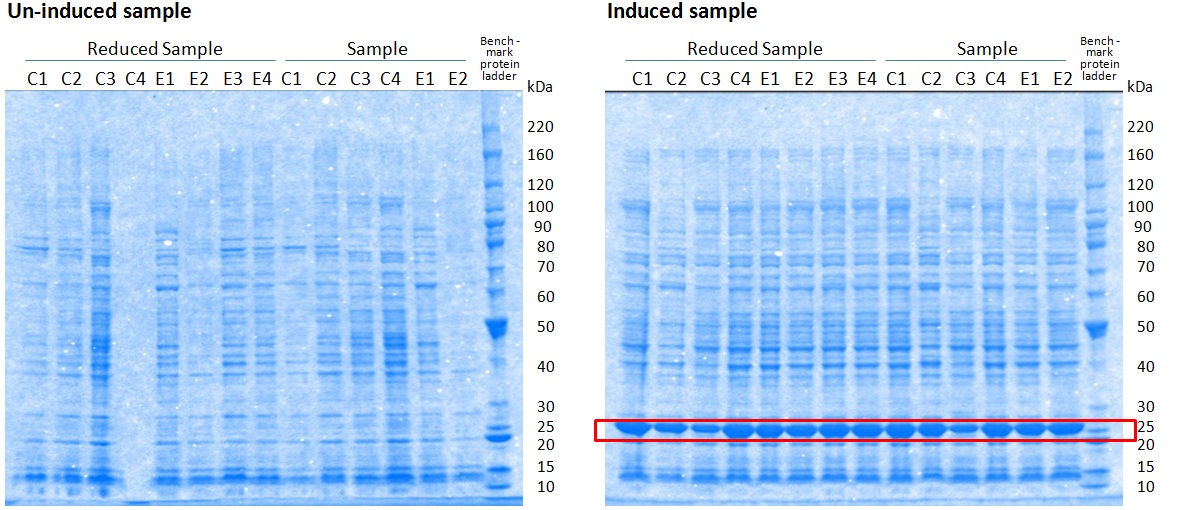
Check the insoluble portion of the lysate culture, trace amount of target protein was detected. It means that the lysation method can be improved to get best yeild.
Uninduced sample Induced sample
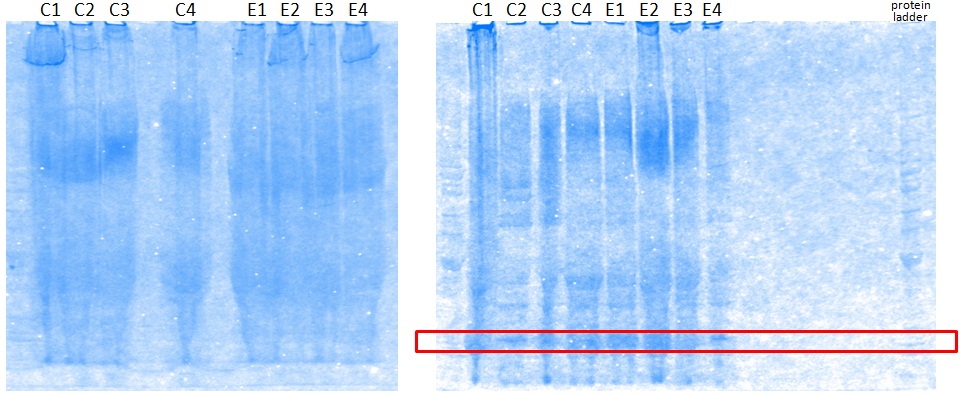
The miniprep pGEX-6p3-PNCA plasmids were digested by EcoR1 and Not1. The target protein size is correct. To further confirm, the plasmids can be sequenced (not now).
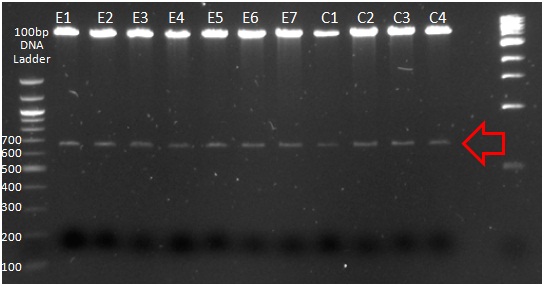
To do:
- Need to repeat Run 3 to keep the consistency of yield batch to batch. By doing so, it will save time in the future experiments.
- Prepare for next step of purification.
- Between experiments, put Eric's references together for new simulation scientist (Will update under folder "Task list for simulation") .
- As Dr Raj mentioned in "Sirtuin Overview", start setting up a kinetic model for sirtuin catalysis based on all the rate constants that have been determined in previous literature using QM/MM and experiments. Will input the related experimental techniques as well (Will update under folder "Sirtuin Overview").
Friday_080213
Starting construct the paper #2. Just put the method of continuous assay and will keep working on it. Second paper template.docx
Need to decide:(a) what journal we would like to consider for submission;
(b) what type of article (communication, full research article) will be;
(c) how much simulation work need to be involved;
(d) what inhibitors will be included;
(e) what experimental data will be incorporated.
RC (8-2): For d,e we should plan to include most of the inhibitors tested so far. Please put info from the wiki and recent experimental results if any into the paper draft.
I have indicated the simulation work involved in a recent email.
Thursday_080113
JMB draft Strategy A with related Table and Figures:
JMB_Strategy_A_072613.docxTable 1_Strategy A_072613.pdfTable 2_070813.pdfTable 3_071013.pdfFig 4_Strategy_A.pdfFig 7_Strategy AD_072613.pdf
JMB draft Strategy B with related Table and Figures:JMB_Strategy_B_072613.docxTable1_Strategy_B.pptxFig 4_071713.pdfFig 7_Strategy B_072613.pdf
JMB draft Strategy D with related Table and Figures:
JMB Strategy D_072613.docxTable 1_Strategy_D.pptxFig 4_070813.pdfFig 7_Strategy D_072613.pdf
Note: The draft contents 10 Figures and 3 Tables, the ones posted here are the changes made according to different strategies.
Friday_050313
Have point-to-point answer Dr. Raj's comments except RC117 and RC118. The related references have been added. The template used for editing is Dr Raj's latest version "Draft of JMB_050213_RC. Draft of JMB_050213_RC_xg.docxRC (5-4): Another possible title for the paper: "Mechanism of inhibition of the human sirtuin deacetylase SIRT3: computational and experimental studies."
XG (5-4): I like this one. It clearly represents the paper (objective and methods).
Tuesday_043013
I have merged my update into Eric's file name as "Draft of JMB_043013_ek.docx". Some of Eric's references were missing due to the use of different version of Word. Plus Eric might add new references which I did not import into the EndNote library. Eric, please confirm this.Draft of JMB_043013_xg.docMonday_042913
Eric, I did not wait for your final merged file and have made some changes based on Dr. Raj's comments under file of "Draft of JMB_042713_rc". Draft of JMB_042913_xg.docDr. Raj, "Draft of JMB_042913_xg" is not final. And the only thing I can do is to merge my changes after Eric's update.
Wednesday_042413
- Literature search for physio-chemical properties of three new molecules
- prepare for solubility tests, solution preparations
- JMB draft update Draft of JMB_042413.docx
Tuesday_042313
JMB draft update based on Dr Raj's comments. Have marked for additional editing. Draft of JMB_042313.docxRC (4-23): I didn't understand whether you have completely removed the inhibition theory section of the discussion. It appears completely crossed out. Also, I believe you have removed the table with the experimental IC50 data for the inhibitors, but the table itself appears to still be there. Are you planning to keep the experimental data only, and not report the computational results, or remove it all?
XG(4-24): The inhibition theory section was replaced by Table 1. The important equations are listed there. Also emphasis couple of reported sirtuin inhibitors followed different inhibition modes. I removed the Table with the experimental IC50 data (it was Table1). I am still not sure if we should only keep the experimental data without reporting simulational results. For the flow of the paper, we should remove it all.
By the way, I just print out the draft, the table with Exp. IC50 data were there. I thought it probably due to the different versions of word I used at PMC-AT. The word on my computer is Word 2007, however, the EndNote was installed on a shared compurter, in which the Word is 2000/2003 version. After I edited references, I need to save the file at 97/2003 compatible form.
Monday_042213
RC's inline comments have been provided, primarily for the computational results and discussion sections, in the attached.Draft of JMB_041813_RC.doc
A few comments have also been provided for the intro. EK is working on an outline of changes to the methods section. He will also address each of the comments listed in the results and discussion sections. He is trying to reuse as much content from presentation slides as possible. RC is in NYC with EK discussing the needed next steps. EK to post his latest changes today. In the computational methods section, RC would recommend having an initial sentence or two indicating that diverse computational biochemistry methods (including some newly developed techniques like customized induced fit) have been used to explain the mechanism of inhibition by NAM and to discover new inhibitors.
XG, please use the "Comments" and "Track Changes" features of MS word going forward, so we can effectively collaborate on revisions.
If possible, I would like the term "Computational" to appear in the title. Also, we may want to use the term "Deacetylase". E.g., "Computational and Experimental Analysis of the Mechanism of Inhibition of the Human SIRT3 Deacetylase." Please provide your thoughts.
We may consider a 3-way discussion later this week if there is sufficient progress. If so, both EK and XG should prepare a list of questions ahead of time.
Thursday_041813
(1) Lastest version of JMB manuscript.Draft of JMB_041813_2.docRC (4-20): This is really hard to read since there is no indication of which parts were recently changed in the latest update (or are these the blue text?). Please provide a more detailed breakdown of changes day-by-day as you make them. Also, there appear to be no changes made by Eric. Regarding Eric's work vis-a-vis this paper, I noticed new LIA results being posted but I thought we were to focus on paper writing without LIA.
XG (4-22): The latest updates are made in blue.
First page: delet the acknowlegement and add Keywords.
Abstract: add one sentense to discribe the three potent inhibitors.
Introduction: Rewrite some of the content and add references.
Results: Delete the old Table 1 and create new one for IC50 and Ki for the 7 molecules (not include NAM and isoNAM). Add respective discriptions.
Discussion: Delete some background and uncompetitve inhibition part. Focus on NAM inhibition and possible mechanism of isoNAM.
Methods: Experiments in lab are done.
Eric will update his part.
(2) List of potential refereesList of Reviewers.docx
(3) Name of associate editor: Dr. Ian Wilson, The Scripps Research Institute. Crystllographic studies of immune recognition and viral pathogenes.
(4) Names of board members to handle our paper (pick one or two among the list):
- Dr. Todd Yeates, UCLA. Molecular, structural and computational biology.
- Dr. David E draper, Johns Hopkins University. Physical Biochemistry.
- Dr. Barry Honig, Columbia University. Biochemistry and molecular biophysics.
Friday_041213
Summary of our discussion on Wednesday about how to finish our first paper.As what Dr Raj suggested, the paper will keep current content.
The work needed Eric to finish are listed below:
(time line added by EK)
• Work on redoing the SIRT3 simulations with the newest SIRT3 crystal structure, PDB:4FVT. (Thurs. 4/18)•Method section should be modified, content need to be shorten, the details are going to supplemental materials. No LIA needed. (Fri 4/19)•Decide which figure should stay (Sir2 work and SIRT3 findings), and combine them if it’s necessary (Fri 4/19)•Decide how to stress Loop Minimization (Mon 4/22)
The work needed Xiangying to finish are listed below:
•Clean up the word and finalize Discussion section.•Write up the findings of three potent hSIRT3 inhibitors (Ex527, Salermide, and AC93253) and add references for smooth transition.•Format paper as JMB required and prepare a cover letter to Editor for paper submission.•Look for few names for list of possible reviewers (normally we will be asked when submit paper).
Saturday_040613
Latest version of ACS presentation04062013.pptxFriday_040513
Latest version of ACS presentation04052013_final.pptx.Thrusday_040413
Update the ACS presentation based on Eric's most recent version "ACS_slides_04.03.2013". 04.04.2013.pptx.
Monday_040113
1. Update the ACS presentation based on Eric's most recent version_04.01.2013. Slides 5 was modified.ACS_slides_04.01.2013.pptx2. Comments on Eric's new slides
- Slide 23_loop minimization: Work from Wolberger’s group has shown that the flexible loop is influenced by NAD+ binding, which can trigger the assembly and disassembly of the C pocket. For example, the structure of Sir2Af2 bound to NAM has ordered flexible loop, but the structure of Sir2Tm bound to NAM has a partially disordered flexible loop from Arg34- to Ser44. Could you prepare a paragraph to describe the different loop conformations between 3GLT and 4FVT? And explain why loop minimization is not necessary? This is part of preparation of our paper.
- (EK 4-3) Added following explaination to slide: Work from Wolberger’s group (2005 Molecular Cell 17, 855) has shown that the flexible loop in Sir2 is influenced by NAD+ binding, which can trigger the assembly and disassembly of the C pocket. For example, the structure of Sir2Af2 bound to NAM has ordered flexible loop, but the structure of Sir2Tm bound to NAM has a partially disordered flexible loop from Arg34- to Ser44. In the above figure, the flexible loop in SIRT3 cocyrstallized with ADPR is disordered (broken brown line). The ADPR, which does not have atoms in the C pocket, does not make as many contacts with the flexible loop as the Carba-NAD structure. The full flexible loop is ordered (defined backbone trace in blue) in the SIRT3 with the Carba-NAD in the productive AC pockets.
- Slides 24 and 25_computational methods: Please provide the detail protocols (Glide, MM-GBSA, induced fit +MMGBSA, and LIA) for the METHOD section of our first paper.
- See attached: docking.methods.docx
- Slide 27_binding energy prediction SIRT3 inhibitor: Ex-527, Salermide, AC93253 are not congeneric series, however, we got good correlation between MM-GBSA and experimental pIC50. Does it mean that ligands with high affinity are important for such a prediction?
- It means that the method worked very well with these three non-congeneric ligands. MM-GBSA may continue to work as well with a larger dataset, but we will not know until we test it. I have lower expectations for R^2 around 0.4, rather than the R2=0.68 and 0.99 shown on the slide with the minimal dataset. The regression will always look better with so few data points.
- Slide 29_SIRT3 loops and residues: Which structure of SIRT1 do you use in left figure? Can you provide a slide in which the sequence alignment of (1) Sir2 and SIRT1, (2) SIRT1 and SIRT3, are included?
- Added sequence alignment for the loop region showing that it is highly conserved. Also added PDB names. In general, I used the PDB file that had the closest thing to NAD+ co-crystallized in the AC pocket. Many of them did not have that available, so I used NAD+ in the AB pocket, or, lastly some intermediate like ADPR. I will discuss this if there is time, as we know the conformation of the loop is affected by NAD+ binding.
- Slide 30_Competitive vs. non-Competitive NAD+ binding: In this slide, SIRT3 structure you used is 3GLT. Are you going to redo the docking by using new SIRT3 cocrystal structure- 4FVT? Are you going to calculate MM-GBSA of Sir2 and SIRT3 for AC, AB binding based on new SIRT3 cocrystal structure?
- Simulations are running now.
Thursday_032813
Update the ACS presentation based on Eric's most recent version_03.24.2013. A slide of Structure/biochemical mechanism of sirtuin deacetylation reaction was added. Some changes were made based on Dr. Raj and Eric's suggestions. 03282013.pptxRC (3-28): The following are missing:
a) clear commentary on why we are seeking SIRT3 inhibitors
XG: Good point. On Slide 5, there is an example: overexpress SIRT3 can induce liver injury. I will make a statement on the slide.
b) how the simulation studies connect to experiment must be stated clearly at the outset of that section (if verbally, notes should be provided). For example, it is not described anywhere how experimental data is used as a training set for the linear MM-GBSA model. In particular, the NAD+/NAM and virtual screening studies are handled very differently (former does not use experimental data for parameter estimation).
XG: Eric mentioned that data from 5 strong inhibitors is necessary for such a training set for the linear MM-GBSA model. For now we have NAM, Ex527, Salermide, and AC93253. Hopefully next week we will get the 3 new compounds so that I can test them before the meeting. The rest of 5 molecules are weak inhibitors, I am not sure if Eric can use them for Linear MM-GBSA model.
RC (3-30): Here I was referring to linear regression of MM-GBSA data not LIA.
EK (4-01): yes, I will add more notes for verbal explanation of the connection between simulation and experiment. Also, Slide "Binding Energy Prediction: SIRT3 Inhibitors" shows trend lines for the 3 experimentally tested inhibitors for SIRT3. I did not put an R^2 value with so few data points. But the trend looks good for MM-GBSA. LIA is not applicable with only 3 data points.
c) the competitive/non-competitive simulation results appear so late, most people will have forgotten their definitions by then
d) there is no detail provided on the new induced fit methods
Still need to add this slide.
XG: What is the status of Project 1? Does it use induced fit + MM-GBSA?
Project 1 (SIRT2 MM-GBSA) jobs continued to run excessively long (multiple days) then die. I have a theory as to why this is happening (the input files with contain the output from Induced Fit, which is not in the proper format for the program). I will run some more simulations today and tomorrow.
e) there is no definition of the terms appearing in the LIA equation
Right, I thought I would briefly explain it verbally. I don't think that we have enough time to go into more details. I added the terms to the slide describing the LIA method.
f) i would like to reiterate that i am looking at the simulation results as work toward the paper draft. we need far more detail to be provided in these slides on all work to date; some will be used as backup slides. This includes a detailed workflow for virtual screening.
RC (3-30): EK, please remember we would like the backup slides for the talk to be sufficient for completion of the paper immediately after the conference. This work should be done prior to the conference, so afterwards it is a straightforward matter to integration into the paper. XG, you should also decide this week what results (including simulation results) you would like to put in the paper.
Wednesday_032713
Skype with Eric and discussed the follwoing issues:1) Eric has added 3 more slides to address the methods used for docking and sumulation study.
(a) Glide XP
(b) MM-GBSA
(c) Induce fit MMGBSA
(d) LIA for Liaison
2) Eric will prepare a slide of two sequence alignments for Sir2 vs. SIRT1 and SIRT1 vs. SIRT3, highlight the highly conserved residues, the residues in the C pocket that make specific contacts with NAD+.
3) Eric will redo the SIRT3 docking study using the new cocrystal structure of SIRT3 with Carba-NAD. Eric will have Glide score and MMGBSA value for molecues we tested in the lab.
4) Eric will prepare a slide show SIRT2 results
5) Discuss with Eric if we need to show Sir2 docking results. For now, since we have SIRT1 and SIRT3 experimental results not Sir2 we will focus on SIRT3.
6) Ask Eric to post the most updated slides and "minutes from discussion with Dr Raj on Tuesday" right after Skype.
Monday_032513
1. 3 compounds from Eric's Best 3 list have been ordered.2. The 1st draft of ACS presentation is posted. I keep experimental and simulation work seperate. I am not sure that's the best way to present. We need to disucss after you all have the first look. Slide 11, I proposed a model. I do not know if Eric's simulation will support it or not. Please try slide show, since there are some animations in the presentation. After Acknowledgement I have some more slides for backup. I will add more when I think it's necessary. 03212013.pptx
Friday_032213
Oder 3 Compounds from Eric's list and run assay.Eric, After you finish all the slides for ACS meeting, would you please provide the Glide-XP scores and MM-GBSA values for the following compounds: Ex-527, Salermide, AC93253, ChemBridge ID 5281077, 4102009, 9147724? Thanks!
Thursday_032113
Please check the below table for details about the availability of compounds in Eric Top10 list. In brief, they are custom synthesis compounds and not available in Sigma-Aldrich.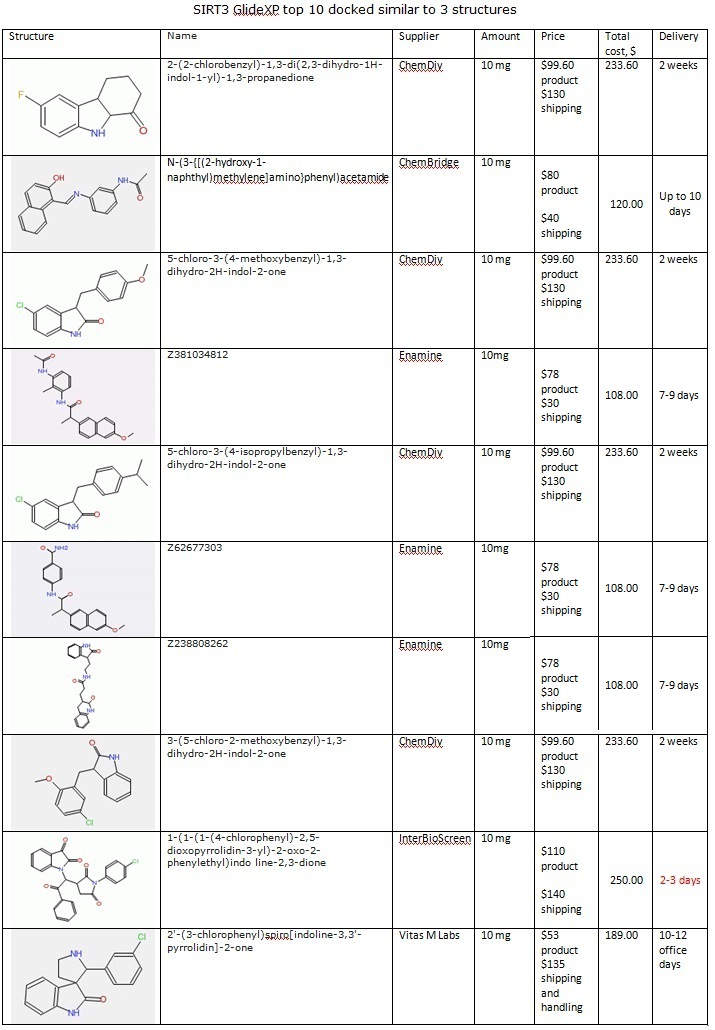
Monday_031813
Discussions between Eric and XG according to Eric's update.Eric:Database queries to www.emolecules.com for the 3 inhibitor molecules you emailed me (Salermide, Indole EX-527, AC-93253) turned up hundreds of similar molecules. However the similarity index sensitivity needed to be reduced from the default 0.8 to 0.7, which resulted in most of the similar molecules being marginally similar. This means that more than just a single side group is different; i.e., some of the core atoms are different. All of them can be used for docking to figure out which one could be used for the assay. All of the molecules are supposedly commercially available from various vendors.
However, because Project 1 is using most of the Schrodinger licenses (for induced fit docking into SIRT2), I think I should pause Project 1 to run a docking job on SIRT3 with these inhibitors. The project 1 computational job has been running full blast and uninterrupted on the computer since Friday, and still has an estimated 48 hours to go.
XG: Please provide the list of all the molecules you mentioned below for your docking study. I need to check their availability.
Eric: there are many molecules (>100). I think I should narrow down the list through docking before you check on availability.
XG: Do you need to prepare the molecules before docking? How long will it take?
XG: How long will take for you to run this docking job on SIRT3, say 100 molecules?
Eric: basic docking on 100 molecules can be done this afternoon if I pause the project 1 simulations. This is basic docking, without the additional induced fit. This could result in more false negatives. But, because of the time constraint, I think this is OK. The docking site will be fitted to find molecules similar to the known inhibitors. The way I will do it is used induced fit on the 3 inhibitors you've already tested to create a model of the docking site to screen with basic GlideXP docking for new molecules from the database of molecules. Hits from this first pass database screen can then be done with induced fit to possibly improve the results, then MM-GBSA done. I am not expecting much correlation between real dG_bind and MM-GBSA or docking scores. This is more of a categorization - which molecules will definitely not bind, vs ones that may bind (one's that you should consider testing)
XG:Basic docking will result more false negatives, what is the percentage of it? You mentioned that “basic docking + induced fit” will provide a list of molecules that may bind. Could you explain that why I shouldn’t expect a better correlation (r2~0.3) between experimental dG-bind and MMGBSA/Glide-XP score? This time, we are going to have enough data points (>12) with strong binding. I assume that you will provide at least 10 molecules for me to test.
RC: I think Eric is saying that the correlations will not be strong since the receptor conformation will not be minimized for each individual molecule, and that he expects stronger correlation on the 2nd induced fit pass on molecules that dock properly.
XG: Does this mean, I should wait the results from his second induced fit pass and select candidate molecules to test? Eric had finished his docking. Eric, please provide the method you used for this docking job, I mean, please confirm that (1) if you have used induced fit to scale down your list (2) is the list ready for me to test?
XG:What is the disadvantages of pausing Project 1? Project 1 is important to us too since that is the way of testing our system with Schrodinger’s new suggestions.
Eric: Pausing project 1 computations for a few hours will just add those few more hours to the computations.
XG: I am willing to see the results of Project 1. I want to know if Schrodinger has better solution. If we cannot obtain r2 around 0.3 at Project 1, what is the point of moving forward for SIRT3 inhibitor screening _ BACK TO POINT 2?
Not a conclusion: Project 1 and Project 2 are equal important to us. The results from Project 1 will provide us a handful knowledge of Sirtuin system. The progress of Project 2 will add a highlight to our ACS presentation. Project 2 has the first priority since XG’s lab work is depended on results of Project 2. However, Project 2 will not be proceeded properly without a good understanding of Project 1.
Dr Raj, we need your advise!
RC: I am ok either way. Eric said the virtual screening jobs take only a few hours. If that is true, it is ok, but it appears they died last night. If there are any further delays (i.e. if this happens again), I would suggest reverting to project 1 until finished.
Friday_031513
Next step on SIRT3 for ACS presentationXG: Provide the molecular structures of Ex527, Salermide, and AC93253 for Eric. (03/15/2013)
Eric: Search for commercial availability of their analogs/derivatives.
Eric: Provide a list of candidate molecules from his docking results.
XG: Order and test in the lab. (~Two weeks)
XG: Calculation of IC50, Ki value. (<2 days)
Eric: Calculation of MM-GBSA and provide correlation between simulation and experimental results.
Dr. Raj, Eric and XG: Discussion and interpretation of valuable data into ACS talk.
ACS presentation
XG: prepare the draft of presentation (need to be finished 03/25/2013, which is two weeks ahead the meeting, for Dr. Raj review)
(1) Background_Aging, Sirtuins, why SIRT3, Why study of inhibition is important
(2) Inhibition_mechanism (briefly go through competitive, noncompetitive)
(3) NAM_ physiological inhibitor, Chemistry of deacetylas reaction
(4) Current method, role of NAM base exchange (future assay will be develop in the lab).
(5) NAM_noncompetitive inhibitor for most of sirtuins, competitive for SIRT3 with experimental data and discussion.
(6) Structure of SIRT3. A, B, and C binding pockets. Features of active and nonactive NAD+, move to simulation work.
(7) Eric please provides your thoughts on this. Please provide all the necessary materials no later than 03/22/2013.
(8) Conclusion and future direction
Long term projects
XG: Develop new assay for testing other sirtuins activities, including receptors, substrate peptide, candidate compounds.
XG: come up a larger database with new molecules and their derivatives for future virtual screening.
XG: Survey of techniques for ligand/protein binding affinity measurement
Thursday_031413
Continue preparing ACS presentations.Wednesday_031313
Construct of ACS presentation:- Backgroud-Aging, Sirtuins, SIRT3, Application, and Significance
- Deacetylation chemistry and mechanism
- Modulator rules: NAM inhibition and IsoNAM activation
- Methods: Experimental and Simulation
- etc
Monday_031113
- The IC50 values for Ex-527, Salermide, and AC93253 are 174.9, 27.2, and 18.2 uM. Under current method, IC50 of NAM for SIRT3 is 36.7 uM. Therefore, Salermide and AC93253 are very good inhibitors of SIRT3. I need to repeat the experiment for another time.
- Will start prepare ACS presentation.
Friday_030813)
The paper Dr Raj just posted is a very interesting paper. Basically it is the continue battle between Pfizer and Sirtris_GSK. Let’s refresh our memory :Ø 2007 Sirtris had published a paper on Nature, in which SRT1720, SRT2183, and SRT1460 were firstly reported as SIRT1 activators with potencies 1,000-fold greater than resveratrol. They are structurally unrelated to resveratrol. Methods they used are high throughput fluorescence polarization for identification and high throughput mass spectrometry (MS) for enzyme activity.
Ø 2010 Pfizer threw out a paper and claimed that SRT1720, SRT2183, SRT1460, and resveratrol are not direct activators of SIRT1. The presence of TAMRA label is necessary for activation SIRT1 by resveratrol along with SRT1460, SRT1720, and SRT2183. To avoid any potential artifacts associated with fluorescently labeled non-native substrates, Pfizer used a native peptide substrate with direct detection and quantification method – HPLC and ELISA.
ØLater 2010 Sirtris addressed that STACs interact directly with SIRT1 and activate deacetylation by an allosteric mechanism.
Ø2013 Sirtris provided more evidence for a common mechanism of SIRT1 regulation by Allosteric activators. STAC1 is SRT2183, and the other compounds are new. Mutagenesis of SIRT1 constructs were performed using a commercial available kit. BIOMOL assay(Fluor de Lys assay), PNC1-OPT assay, Oacetyl ADP ribose MS assay, and continuous assay were used for their kinetic measurements. ITC for protein-ligand binding, CD for determination of protein/peptide confirmation. It is very useful to review all the experimental methods for our own use. Models of assisted allosteric activation (AAA) of SIRT1 by small molecules were proposed.
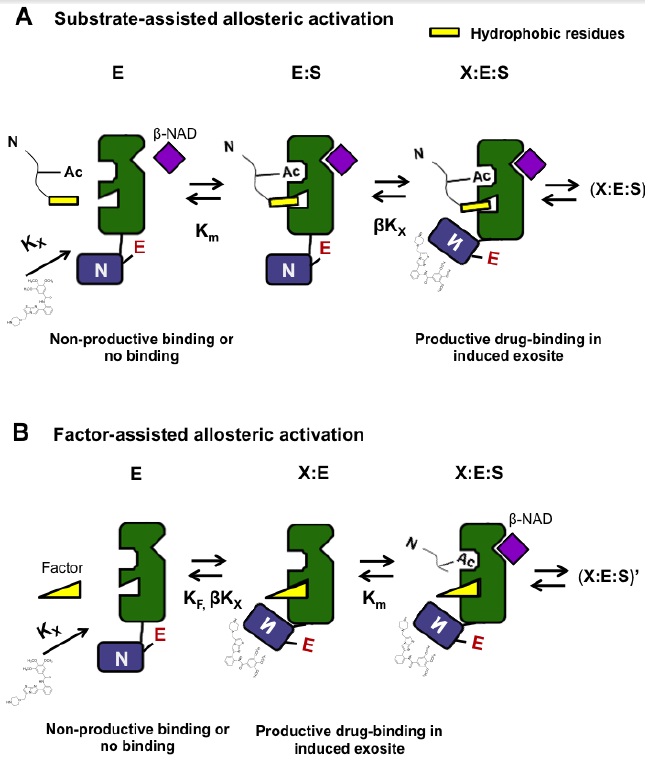
A) substrate-assisted allosteric activation and B) factor-assisted allostericactivation.
Yellow=hydrophobic residues C-terminal to the acetylated lysine.
Red E =E230, whose negative charge may interact with a positively-charged amino acid in SIRT1 to facilitate alllosteric activation.
E=enzyme;
S=substrate;
X=STAC;
X:E:S=activator:enzyme:substrate complex;
KX=activator equilibrium constant;
KM=Michaelisconstant;
beta-KX=activator equilibrium constant in the presence of a docked substrate/factor;
KF=factor equilibrium constant.
Tuesday(030513) - Monday (031113)
Test molecules in the lab. Will update the progress and results every so often.Monday_030413
1. Precise calculations for testing new inhibitors.2. Studies and preparation of stock solutions in the lab (solubilities of 3 compounds)
- Salermide is soluble in organic solvents (DMSO, dimethyl formamide_DMF). The solubility of salermide in these solvents is approximately 30 mg/ml.
- For Ex-527, about 20 mg/ml in DMSO & DMF.
- Ex-527 and Salermide are sparingly soluble in aqueous buffers. To prepare such a solution for maximum solubility in aqueous buffer, the compound should first be dissolved in organic solvnet (DMSO/DMF). Then diluted with the aqueous buffer like PBS (pH=7.2).
- The aqueous solution is not good for more than one day. Need to prepare right before experiments.
- AC-93253 is not soluble in water. Solubility of it is ~10 mg/ml in DMSO.
- Will use DMSO as the negative control in the experiment. The total of addition of DMSO is 5 ul in 100 ul of reaction solution.
Friday_030113
*Chemicals ordered on 03272013 are expected to be in the lab on Monday.*Plan experiments for new inhibitors
*Start to construct ACS presentation. The cover will look like the following figure, not the final version.

*Radioactive labeled reagents are involved in all of the current published methods for detection of NAM exchange rate.
Microplate filtration assay for NAM release from NAD using a boronic acid resin. McDonagh T et al. Methods 36 (2005) 346-350.
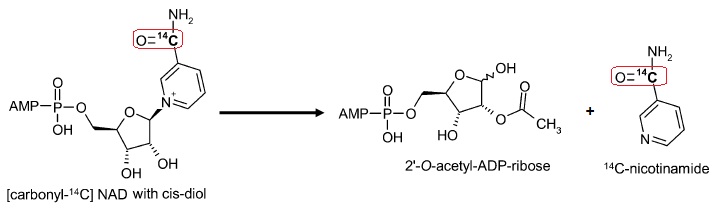
Sir2 Regulation by NAM results from switching between base exchange and deacetylation chemistry. Sauve AA and schramm VL. Biochemistry 42(2003)9249-9256.
Mechanism of NAM inhibition and Transglycosidation by Sir2 Hisotone/Protein deacetylases. Jackson MD et al. J Biol Chem. 278 (2003)50985-50998.
Chemical activation of Sir2 dependent silencing by relief of NAM inhibition. Sauve AA et al. Mol. Cell 17 (2005) 595-601.
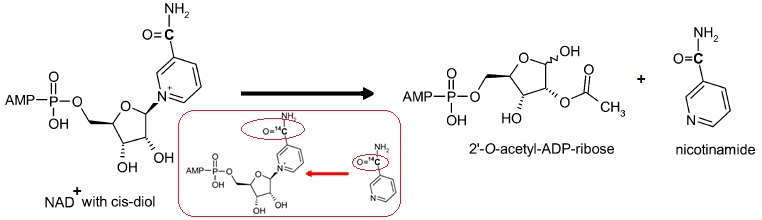
Thursday_022813
1. Some modifications were made in JMB manuscript, mainly at Discussion section in green. Outline of JMB_022813.docFew groups have been reported that NAM can react to regenerate acetyllysine and NAD+ in a nicotinamide exchange reaction, in which the imidate intermediate is emptied during normal steady-state turnover, directing NAM inhibition of deacetylation.By using [carbonyl-14C] nicotinamide the Base Exchange reaction for Sir2 was extensively studied (Scheme 2).
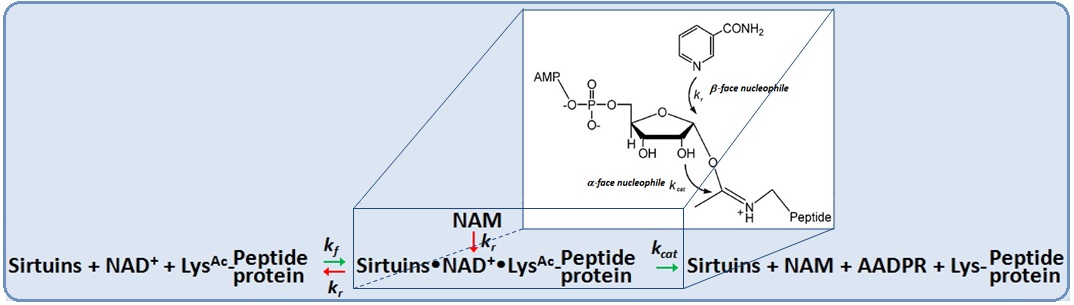
Scheme 2.The competitive nucleophilic attacks on the Sir2 intermediate occur from both stereochemical faces.
The reactivity between Base Exchange and deacetylation reactions occurs when NAM is cound. This competition partitions the intermediate forward and backward to provide inhibition of deacetylation. The independence of the deacetylation and base exchange reactions establishes that base exchange is a b-face nucleophile process, whereas deacetylation is an a-face nucleophilic process.NAM partition ratios are controlled by the relative rates of nucleophilic chemistry at the b-face versus the a-face of the intermediate. The exchange and deacetylation reactions share the intermediate forming step, and the ratio is determined by the chemical processes. It seems likely that unreactive isosteres of NAM that interfere with NAM binding could derepress sirtuin enzymatic activity and increase sirtuin function
2. Discuss with Eric about simulation work.
- XG: Our current priority is to figure out what and why cause the differences between our results and literature data. It might be some bugs in Schrodinger software.
- XG: We need to run other 3 - 4 sets of data. If the problem is still there, we make a strong case of getting attention from Schrodinger to assist you and fix it.
3. What we can do is
- XG: Make a list of simulations you picked from published paper, in which MM-GBSA / MM-PBSA were applied for binding affinity calculation. It does not need to be sirtuin related.
- XG: Run such a set of data and post it on a daily basis.
- XG: At the meantime, start reaching out Schrodinger Tech Support because it takes time for them to understand our problem and get the right person.
RC: Early next week we should continue to run more data sets while waiting to hear back from schrodinger.
Wednesday_022713
1. Chembridge Corporation, San Diego, USA--for customized molecules synthesis2. Notes from paper posted yesterday
- The 10 thiobarbiturates and barbiturate, as training set, are taken from the following paper.
Can anybody get this paper please?
- 14 novel thiobarbiturates were tested by using a homogeneous fluorescent deacetylase assay (similar to Fluor-de-Lys kit). All compounds gave IC50 values in the range between 1-6uM, which show a good potency for SIRT2 inhibition.
- The most favorable interaction field is observed in the acetyl lysine binding channel of SIRT2.
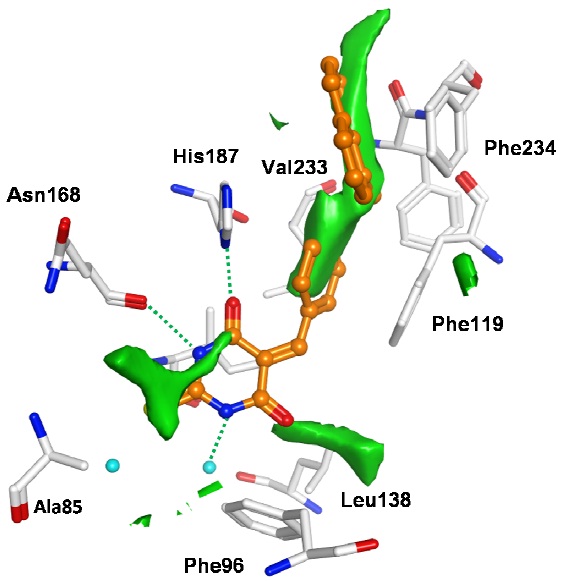
- The supplemental materials with detailed computational methods and experimental procedures are attached. Supp_Binding free energy calculations and biologicaltesting of novel thiobarbiturates as inhibitors_Uciechowska_2012.pdf
Tuesday_022613
1. Cell lysate buffers were prepared.2. The cell culture was pelleted and washed by 1XPBS buffer (4oC) and stored in -80oC for further use.
3. List of 6 molecules has been generated.02262013 inhibitor need to be tested in the lab.docx They are reported as potent SIRT1/SIRT3 inhibitors. Three of them (Salermide, EX-527, and AC-93253 with IC50 of ~50, 49, and 24.6uM respectively) are commercially available.
4. Have sent the request for ordering the afformentioned chemicals for lab test.
5. Literature update. Binding free energy calculations and biologicaltesting of novel thiobarbiturates as inhibitors_Uciechowska_2012.pdf
A multi-step virtual screening was carried out in order to identify inhibitors of human NAD dependent deacetylase sirtuin 2 (Sirt2). Molecular mechanics Poisson–Boltzmann surface area
(MM-PB/SA) and linear interaction energy (LIE) calculations were carried out on a training set of ten recently identified Sirt2 inhibitors. The docking scores did not reproduce the
relative binding free energies estimated from the in vitro data, while the LIE and the MM-PB/SA data were found to be in good agreement with the experimental data for the ten inhibitors. Both binding free energy methods were successful in predicting the activity of 14 novel identified thiobarbiturates and led to Sirt2 inhibitors that are ten-fold more active than those from the training set. The provided data obtained by the combination of docking and MD-based binding free energy calculations show the performance of the approach for predicting the binding free energy of novel sirtuin inhibitors.
Monday_022513
1. Prepare the lysis buffers A, and B2. Pellet and sonicate cells to get whole protein from cell lysates.
3. Update manuscript. Additions are in green. Outline of JMB_022213.doc
4. Summary of meeting with Dr Raj (02/25/2013)
v Repeat reported simulation results _first priority
- Thrombin regression or choose another set of data to test.
(b) Can we reproduce the results from "studies on Splitomicin derivatives paper_J Med Chem 51 (2008) 1203-1213."
- Dock NAM into C pocket of ySir2 (2H4F) to obtain Glide-XP score. The Ki (NAM) for ySir2 was reported as 115 uM with Delta G of binding -2.313 kcal/mol.
- If it works, Sir2 assay will be developed and applied in lab for small molecule screening
- Need to find few more such molecules for Eric to dock to ySir2
v Review paper titled as “Structure-activity studies on Suramin analogues as inhibitors of NAD-dependent histone deacetylasese. ChemMedChem 2(2007)1419.”
- Need to develop similar figure as Figure 1C (ChemMedChem 2(2007)1419) in our manuscript.
- No comparison of experimental data and simulation results.
- Structure-activity studies on Suramin analogues as inhibitors of NAD-dependent histone deacetylasese. ChemMedChem 2(2007)1419.
- Structure-activity studies on Splitomicin derivatives as sirtuin inhibitors and computational prediction of binding mode. J Med. Chem. 51 (2008) 1203.
- Few paper Eric mentioned previously (docking studies on sirtuin inhibitor discovery).
- Finish up the paper with the tested potent inhibitor(s)
- We will report simulation results if Glide XP/MM-GBSA scores match the experimental data.
- Otherwise, experimental data will be included in the paper and mention of ongoing simulation work.
- Figures: filter down the pictures, group them if necessary, edit figure legend.—the final figures will be used for ACS presentation as well.
- Method, Results, and Discussions need to be finalized.
Friday_022213
RC: I am trying to understand what is considered a drug candidate for sirtuin inhibition given its Ki. I.e., what binding affinities are we aiming for and is it important to accurately predict the binding affinities of the types of molecules we have assayed so far (or are they orders of magnitude too weak in binding, so we do not need to accurately predict such weak bindingi affinities). Are any of these considered drug candidates?XG: NAM is the endogenous inhibitor of sirtuins. The Ki (NAM) for SIRT1 is about 50 uM [Kiviranta PH et al, J Med Chem 52 (2009) 2153], 100 uM for SIRT2 [Tervo AJ et al, J Med Chem 47 (2004)6292], 130 uM for yHst2 [Denu JM et al, JBC 279(2004) 40122], and 11.6 uM for hSIRT3 (current work), given their Delta G of binding are -2.525(SIRT1), -2.348 (SIRT2), -2.281(yHst2), and -2.897 kcal/mol (SIRT3) respectively. In general, the potent drug candidates for sirtuin inhibition we are aiming for should have lower Delta G of binding compare to NAM. Taking account of toxicity, synthesis small molecules must be potent enough to replace the natural regulation product, like NAM. Therefore, the molecules tested in the lab are not considered as drug candidates.
It is not clear to me, in our case, if the orders of magnitude too weak in binding make it impossible to get accurate value. I agree that accurate prediction of binding affinities of inhibitors is important. Taking consideration of not having the right starting co-crystal structure, I recalled that we had done some docking studies on 2H4F. Do we have Glide-XP score of NAM for ySir2? The Ki (NAM) for ySir2 was reported as 115 uM with Delta G of binding -2.313 kcal/mol, can make comparison?
RC: I would like to discuss this with you on Mon or Tues as soon as Eric is able to resolve the discrepancy between his results and the published thrombin literature.
Also, I saw results in the past of inhibitor docking to sirtuins. What pocket did these dock in?XG, Please comment on this. Can you remind me of what molecules these prior sirtuin docking studies looked at?
Pyridine, splitomicin, cambinol, suramin, sirtinol, salermide, indole, tenovin, AGK2 and their analogues (or derivates) were the major focus of sirtuin inhibitor investments. Among them, splitomicin, sirtinol, salermide thiobarbiturate with their analogues were docked into NAM binding site of sirtuins (mainly hSIRT2). Some potent sirtuin inhibitors can bind to B, C pockets and peptide substrate binding site. Dr. Manfred Jung’s group has published few papers on screening sirtuin inhibitors using computational prediction combined with the measurement of inhibitors IC50. Inhibitor docking, virtual screening, molecular dynamics and thermodynamic computations were involved in their study. Specifically, H2O molecules were used for docking study and energy contributions to the free energy of binding were provided as well.
Structure-activity studies on Suramin analogues as inhibitors of NAD-dependent histone deacetylasese. ChemMedChem 2(2007)1419. Structure activity studies on suramin_2007_Trapp.pdf
RC: Eric should have been looking at these papers as well. They are directly relevant to his work and not being aware of them as we write our paper is risky. We need to position our work vis-a-vis this prior art.
- Suramin inhibits SIRT5 activity (IC50=22mM) by binding into the B- and C-pockets of the NAD+-binding site as well as to the substrate-binding site.
- Suramin acts as a linker molecule resulting in dimerization of SIRT5.
- Detailed structure–activity relationships on splitomicin derivatives and their inhibition of recombinant SIRT2.
- Ligand docking followed by molecular mechanics Poisson–Boltzmann/surface area (MM-PBSA) calculations
- Protein-based virtual screening resulted in the identification of a novel Sirt2 inhibitor chemotype.
Thursday_022113
1. Overproduced cell cultures will be midiprep and store into -20oC freezer for sequencing.2. Design and order the premir pairs for sequencing.
3. Overproduced cell cultures wil be divided into two protions as 1)control, 2) induced with IPTG.
4. Summary for reported SIRT1 inhibitors with IC50 not limited for NAM binding site. Known SIRT1 inhibitors.docx
5. Literature search for new assay_NAM exchange (not finished)
RC: XG, would you please provide a link to or summary of the method you used to convert IC_50 to K_i.
XG: http://botdb.abcc.ncifcrf.gov/toxin/kiConverter.jsp
RC: Please confirm that that the transformation from IC50 to Ki that you used is linear and provide a short write up on it for the paper if you have not already done so, if it is in fact necessary for obtaining Ki's from the experimental data.
XG: The transformation from IC50 to Ki is linear. Experimental conditions are designed to fulfill the following general assumptions (a) the NAD+- and inhibitor- binding reactions are reversible (b) the enzymatic reaction is one-to-one stoichiometry (c) no enzyme autocleavage occurs (d) concentrations of substrate and inhibitor used are in excess of the enzyme concentration. Hawse WF et al from Dr. Wolberger's group used same equation to obtain Ki value from IC50. (Structural insights into intermediate steps in the Sir2 deacetylation reaction. Structure 16 (2008) 1368-1377.)
6. A short discription of IC50-to-Ki calculation for paper.
Monday_021813
1. 6 single colonies were inoculated in 3 ml of LB/Ampicilin solution overnight for OD~0.62. 1 ml of above solution will be prepared as glycerol stock and stored in -70oC.
3. overproduce above solution in 5 ml and midiprep for sequencing.
4. Manuscript modification (ongoing)_add discussion based on paper "AA Sauve and VL Schramm. Biochemistry 42 (2003) 9249-9256.".
5. Literature search for new assay_NAM exchange.
6. Literature highlight: The first structural study on Sirtuin-activator. RC: Yes, I believe saw this paper. Modeling this may be difficult.
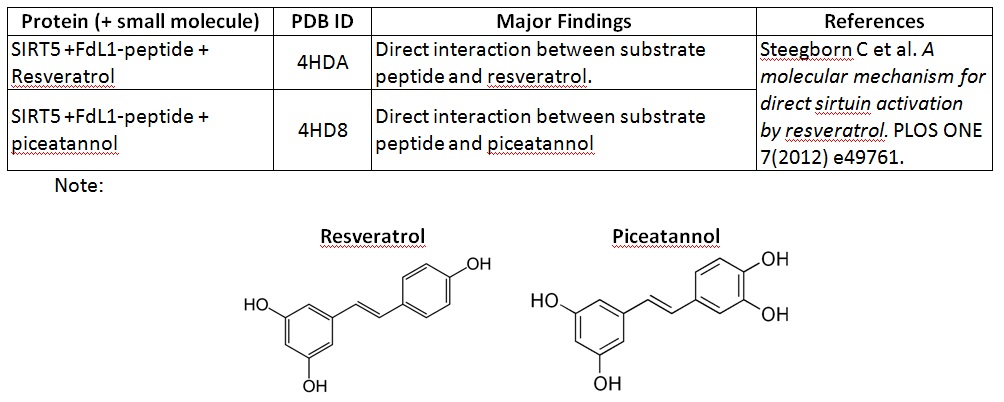
- Resveratrol (3,4',5-trans-trihydroxystilbene), a naturally occurring stilbene, is considered to have a number of beneficial effects, including anticancer, anti-aethrogenic, anti-oxidative, anti-inflammatory, anti-microbial and estrogenic activity.
- Piceatannol (3, 3', 4, 5'-trans-trihydroxystilbene), a naturally occurring hydroxylated analogue of resveratrol, is less studied than resveratrol but displays a wide spectrum of biological activity. Piceatannol has been found in various plants, including grapes, passion fruit, white tea, and Japanese knotweed. Besides antioxidative effects, piceatannol exhibits potential anticancer properties as suggested by its ability to suppress proliferation of a wide variety of tumor cells, including leukemia, lymphoma; cancers of the breast, prostate, colon and melanoma.
- Model for the regulation of Sirtuins by resveratrol-like compounds. After binding of the substrate polypeptide the small molecule attaches to a ‘‘docking patch’’ (DP). It induces ordering of an ‘‘adaptable loop’’ (AL), leading to closure of the peptide exit and stabilization of the enzyme/ substrate complex. Depending on the fit between substrate and small-molecule, the substrate is properly oriented in the active site (AS; e.g. Sirt5/ Cytochrome c/resveratrol) or adopts a non-productive conformation (e.g. Sirt3/GDH/resveratrol), leading to stimulation or inhibition of turnover, respectively. After deacetylation, the activator dissociates, opening the peptide exit for product release.
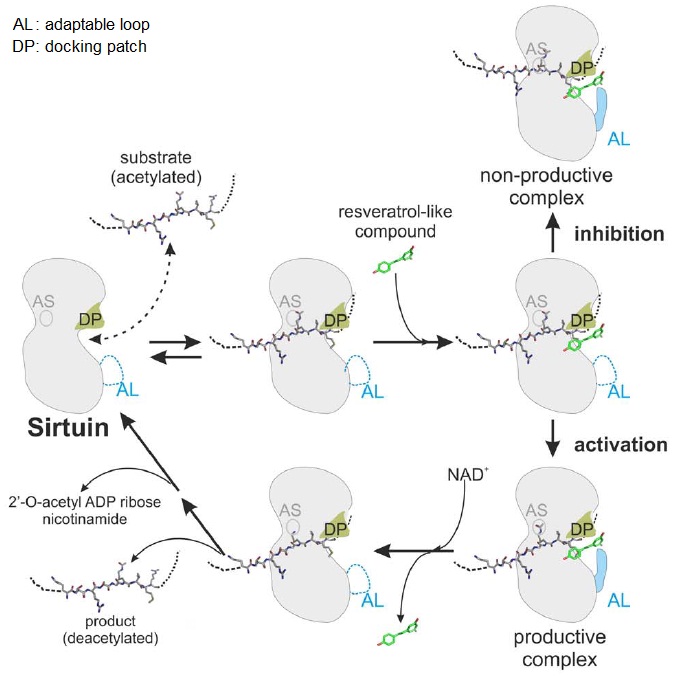
Thursday_021413
1.Parental pGEX-6p-3 and pGEX-6P3-PncA plasmid DNA have transformed into E Coli BL 21 strain.2.The above transforms were plated on LB argar w/ampicilin 37oC, overnight.
3.Inoculate selected single colonies at 37oc overnight.
4.Reference highlight: AA Sauve and VL Schramm. Biochemistry 42 (2003) 9249-9256.
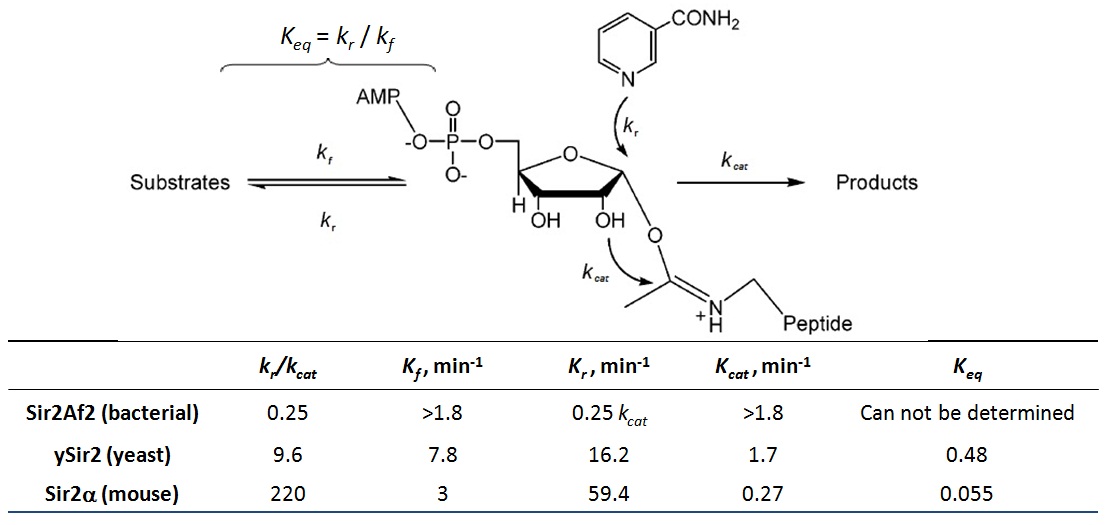
The kr/kcat ratio is calculated by the kcat(exchange)/kinh(deacetylation) ratio.
For the yeast and mouse enzymes, the value of kf is determined by kcat(exchange) + kinh(deacetylation).
The kcat value is determined from kcat(deacetylation).
The value of kr is computed from the determined kr/kcat ratio and the value of kcat.
The equilibrium constant is calculated from the rate constants kfand kr.
Note: kcat(deacetylation) is the rate of the deacetylation reaction for the enzyme in the absence of added nicotinamide.
kcat(exchange) is determined from the saturation curves for exchange (exchange reaction rates vs. nicotinamide concentration).
kinh(deacetylation) is the residual deacetylation rate in the presence of 2 mMnicotinamide.
RC: XG, yes, we discussed before how obtaining the Keq for NAD binding requires kr and that it cannot be obtained from our current experiments. Is the kr above that for
NAD binding to the active site? Is Keq that for NAD binding? Are you indicating that an assay capable of looking at base exchange kinetics can provide us with kr? By posting these
literature data are you following up on our earlier discussions and indicating that you would like to set up such an assay (for a future paper)? Or are you suggesting that we should incorporate
some more discussion on these points in our current paper? That would seem like a good idea.
XG: Kr is for produced NAM binding to active site. Keq is for NAD binding. There are couple of assays in which the base exchange kinetics can be measured, [carbonyl-14C]nicotinamide, HPLC.. I would like to incorporate some discussion on these points in our current paper. I would like to setup an assay for our future paper by not using radioactive reagents (need more literature reasearch on it).
Tuesday_021213
1. List of sirtuin inhibitors of NAM binding site. Sirtuins inhibitors for NAM binding site_02122013.docx2. JMB manuscript reformates.
3. Paper/pencil calculation based on couple of reported experiments.
4. Setup and preparation for unfinished continuous assay.
Thursday_020713
1. Kinetic studies revealed that the acetylated lysine binds to the enzyme prior to NAD+ and that nicotinamide is the first product released.--------Borra, M, Langer, M, Slama, J, Denu, J. Biochemistry 43 (2004) 9877.------ In our reaction system, ordered sequential mechanism can be applied, see the following Scheme:
However, under current Flour de Lys method, the base exchange rate can not be quantitated. Km, Kcat values are overall effect.
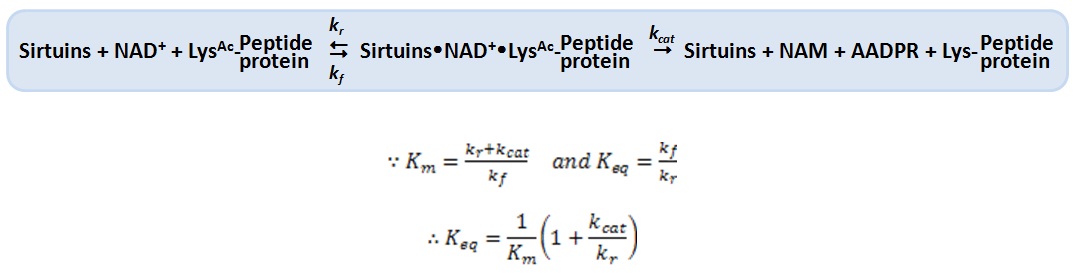
2. Km, Kcat values for hSIRT3 and hSIRT1 using current method are listed

3.The IC50 and Ki value for 6 molecules:
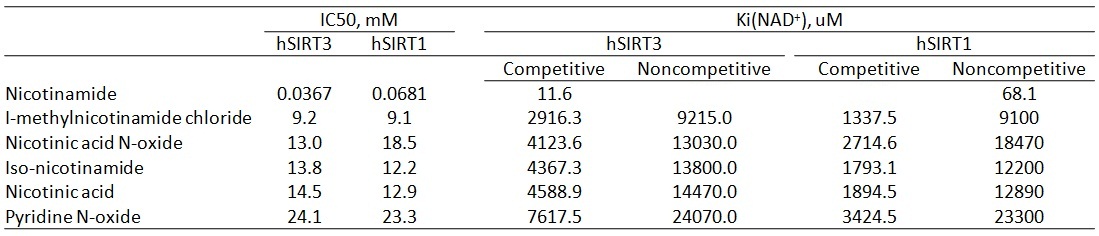
Note: The Ki value is calculated from IC50. The Ki value for both competitive and noncompetitive inhibition are listed for comparison.
Except NAM the rest 5 molecules are not strong inhibitors for either hSIRT1 or hSIRT3. In order to report these data, we need to have at least one molecule which has better profermance.
RC: Are you referring to better performance so we can find an effective inhibitor or in order to run a linear regression to predict binding affinities of new inhibitors computationally? The two would seem distinct.
XG: Based on experimental results, the 5 small molecules I tested have very little inhibition effect on hSIRT3 activity. We want to report at lease one compound (not necessary to be new), which is an effective inhibitor of hSIRT3 by comparing with NAM (IC50 and Ki values fall in same order of magnitude). In another word, experimental resultes tell us these 5 molecules are not good. No matter what kind of computational methods we use, there is no excitement to report these 5 weak inhibitors of hSIRT3 in our paper. However, Eric can test his methods and see if his numbers matches my results.
RC: If we are able to use this data to predict binding affinities of new inhibitors computationally, that would be a milestone in itself. However, due to the concern immediately below, I agree we may need more tightly binding inhibitors. However, we can always test our prediction accuracy with new inhibitors, even if they are weakly binding, to check accuracy.
RC: Are you indicating that if the molecules do not dock sufficiently tightly to the C pocket, we cannot accurately compute their Ki's using the Michaelis-Menten model since the latter molecule omits the other important kinetic steps above? I recall we discussed this earlier.
XG: Yes. I do concern that we might not accurate compute the Ki's using Michaelis-Menten model because the aformentioned important kinetic steps are omited. To prove it, we need a bigger amount of smaples to dock and test.
RC: I agree with this. We should be concerned about the accuracy of the Ki model when we compare experimental to computational binding affinity data. We will see how well they correlate shortly once Eric finishes his regressions.
The chemicals, which are available in pmc-at lab, are listed as following (it was posted on WIKI at 01/07/13). Most of them were reported that can be docked into nicotinamide binding site.
RC: Who reported that these dock into the NAM binding site? All of them?
XG: These compounds are available in our lab. They are reported as SIRTUIN inhibitors, but not only dock into NAM binding site. Like, Suramin sodium, it was reported to inhibit SIRT5 by binding into the B and C pockets of the NAD+ binding site as well as the substrate-binding site. It acts as a linker molecule resulting in dimerization of SIRT5.
I can narrow down my collection of sirtuin inhibitor library for only NAM binding site.
RC: Yes please do so.
REC
4. Discuss with Dr. Raj and Eric about
(a) if we need to test more small molecules, and which of them are picked.
RC: Please let me know if you would like to discuss.
XG: As what I mentioned above, we can report these 5 molecules, but we would like to include one effective inhibitor as well. To do so, it will be nice to change the order of procedure. I have prepared a list of reported inhibitors of ySir2, SIRT1, Hst2... last year. I have just found another paper in which 10 compounds were tested for inhibition effect of hSIRT1/hSIRT2/hSIRT3. It will take me a day or two to put them together as a library. I would like Eric to screen them first, then I run the experiments to test if they are good. It will save some time for me to finish the continuous assay and experimental cost.
RC: Yes I agree and I mentioned this last week. This is why I indicated we needed Eric to start testing new molecules even before you finished your experiments, over the last two weeks. Please post the list asap.
(b) if we need to have a experiment to test if any of them has similar behavior as iso-NAM-------Sirtuin activator by relief of NAM inhibition.
RC: This is of lower priority since the focus of this paper is to predict binding affinities of inhibitors - we do not have the ability to determine the other rate constants above which
are important for activation. We will do that later.
XG: OK.
5. Literature search for more novel inhibitors to add to the list.
6. Edit our JMB manuscript based on new results.
Monday_020413
Will add the IC50, Ki for them
Will list the Km, Kcat for hSIRT3 (NAD+_).
Thursday_01/31/13
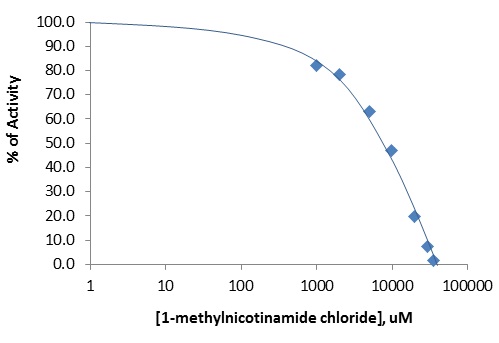
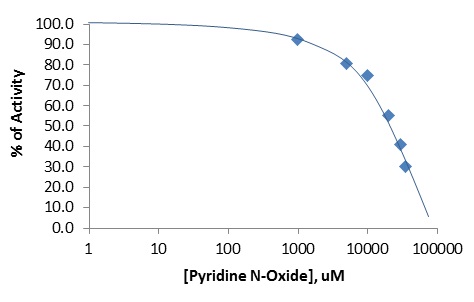
Finish the experiments by early next week for both hSIRT1 and hSIRT3. Calculations are on the way.
Thursday_01/24/13
1. Sirtuin kinetic parameters overview (method for kinetic study is included).Summary of Sirtuin Kinetic Parameters 2013 update.pdf2. Reference for IC50-to-Ki Calculation.supp_Structural insights into intermediate steps in the Sir2_2008_Hawse.pdfStructural insights into intermediate steps in the Sir2_2008_Hawse.pdf
3. Need more time on finishing SIRT1/SIRT3 inhibition by small molecules experiments.
Monday_01/21/13
1. Sirtuin structure overview Summary of Sirtuin Structures 2013 update.pdf2. Sirtuin kinetic parameter overview_not finish
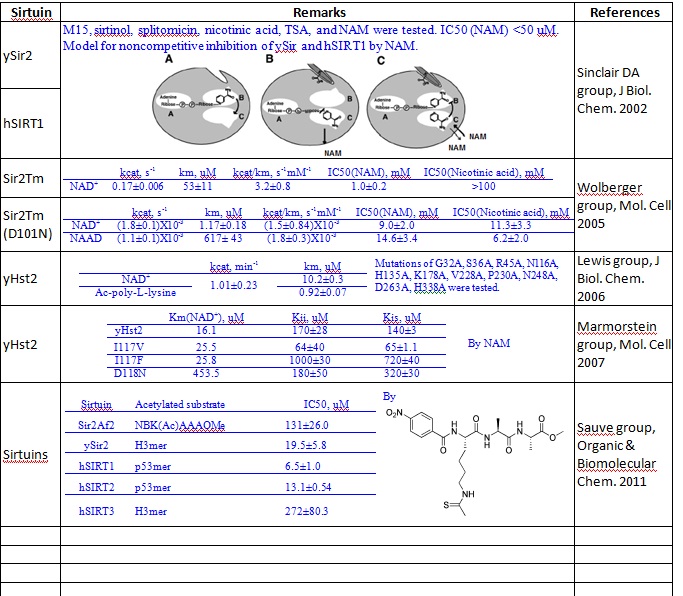
3.Setup and test pyridine N-oxide, 1-methylnicotinamide chloride, nicotinic acid N-oxide, and nicotinic acid for inhibition (IC50, Ki) of hSIRT3.
Thursday_01/17/13
1) Experimental conditions have been setup ([inhibitor]= 5 uM - 5 mM) and pyridine N-oxide, 1-methylnicotinamide chloride and nicotinic acid N-oxide have been tested for inhibition of hSIRT1. Will run another independent experiment to obtain IC50 and Ki.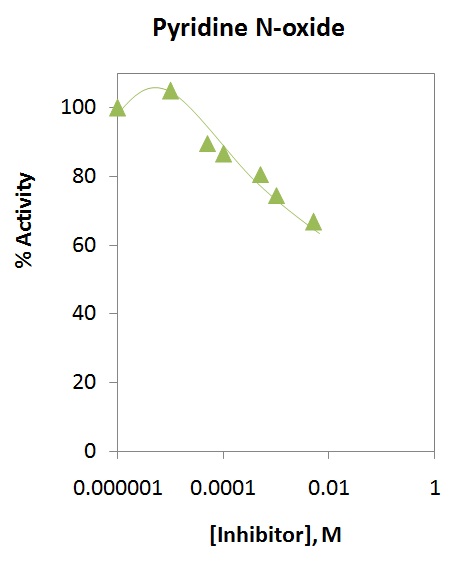
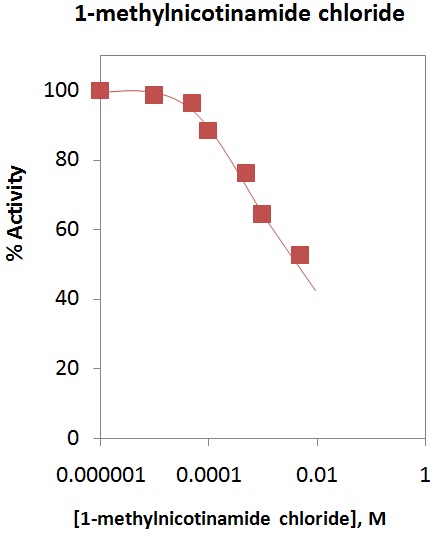
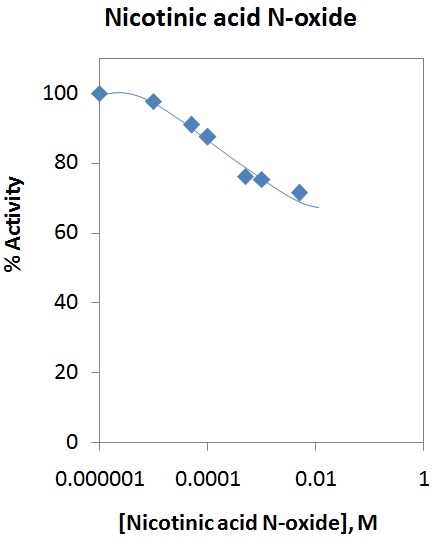
2) Will test the aformentioned molecules and nicotinic acid for inhibition (IC50, Ki) of hSIRT3.
3) Half way through the calculation of Km, Vmax, Kcat under appropriate units.
4) 70% done on literature review of the Km and Ki for NAM for Sirtuins.
RC (1-15): XG, are you also working on reporting kcat, Km for Sir2/SIRT3 with respect to NAD+ in the appropriate units?
XG: Yes, I am working on them too.
RC: Regarding literature review, there were a couple of papers/reviews that discussed how the Km and Ki for NAM for Sir2 are of similar magnitudes, and
how this was predicted based on the based on the potency of NAM as an inhibitor. I may be misquoting this but we should find these values and references if possible
and compare the values for SIRT3 once you have them in correct units. Not a first priority, I am just noting it so we do not forget.
XG: A good point. I have few paper on hand, will make a table for comparing and discussion.
Monday_01/14/13
1)The concentrations of 1-methylnicotinamide, pyridine n-oxide, and nicotinic acid n-oxide were screened for SIRT1/SIRT3 kinetic assay.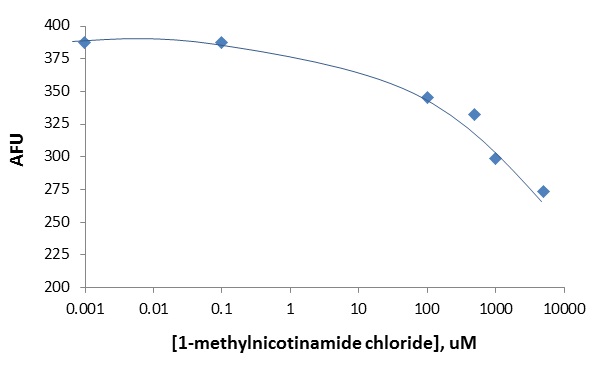
Two issues need to be evaluate further (a) solubility of the reagent to make high concentrated solution, how far it can go
(b) if the activity of SIRT1/SIRT3 can be fully inhibited by aformentioned molecules. On the other word if the reagents are only partially inhibit
SIRT1/SIRT3 under corrent experimental condition.
2) The ki value will be obtained when I figure out (a) and (b).
Thursday_01/10/13
1) Have done first set of experiments for screening the reasonable range of inhibitor concentration and continue working on the jobs listed below.3) Skype with Eric Friday.
Monday_01/07/13
1) The protocol of kinetic assay for inhibition experiments of getting Ki for the 6 reported (our JMB manuscript) inhibitor of SIRT3 has been generated.2) Prepare the stock and working solutions for pyridin N-oxide, 1-methylnicotinamide chloride, and nicotinic acid N-oxide under concentration range of 0-50mM.
3) Perform a titration experiment and provide an estimate time frame.
4) Continue literature searching to get necessary reported kinetic parameters for discussion
5) The list of inhibitors which are available in lab_ Most of the reported molecules can be docked into nicotinamide binding site.
Thursday_01/03/2013
SIRTUIN BIOLOGY DEVELOPMENTS UPDATESirtuin activators
SRT1720 improves survival and healthspan of obese mice, Minor, R.K. et al. Scientific Reports 1:70, 1-10.
SRT1720 improves survival and healthspan of obese mice_Minor_2011.pdfsupp_SRT1720 improves survival and healthspan of obese mice_Minor_2011.pdf
- SRT1720 extends lifespan and confers hepatic and pancreatic protection in obese mice.
- SRT1720 improves body composition, glucose homeostasis and metabolism in mice on a high-fat diet. These results consistent with the effects of resveratrol, which was also found to reduce cardiovascular and liver pathologies in conjunction with increased survival.
- Similar as resveratrol, SRT1720 suppresses hepatic apoptosis and normalizes gene expression in obese mice.
- Both resveratrol and SRT1720 treatment increase mitochondrial biogenesis.
- Structures of resveratrol and SRT1720
- The debate as to whether SRT1720 and resveratrol are direct activators of Sirt1 has been around for a while.
- The current In vitro results show that SRT1720 promotes Sirt1 deacetylase activity in vitro towards p53.
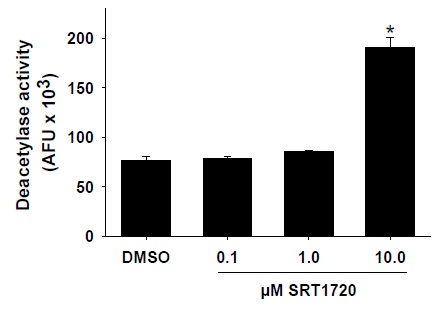
- However, the present in vivo effects by SRT1720 on healthspan or lifespan do not distinguish between direct and indirect Sirt1 activation.
Sirtris patents overview
In 2012, 4 patents were applied by Sirtris Pharmaceuticals, Inc., which are
The details of the patents will be discussed other where.
Sirtris pipeline overview

SIRT3 and SIRT6 are regulators of longevity
Although SIRT1 is the most well studied sirtuin genes that were reported to extend lifespan in lower organisms, the first sirtuin to be associated with longevity in a human population was SIRT3. A variable enhancer region for SIRT3 was identified to show that individuals carrying the alleles with the lowest enhancer activity were the least likely to survive to advanced ages.
SIRT3 mediates the induction of antioxidant defenses and metabolic adaptations during caloric restriction. Caloric restriction reduces oxidative damage in the brain and liver, prevents the loss of neurons and hair cells from the inner ear, and dramatically attenuates age-related hearing loss in mice, and each of these effects requires SIRT3.
SIRT3 is also induced by caloric restriction in white adipose tissue, brown adipose tissue and skeletal muscle, and mediates adaptive changes in hepatic metabolism, including the upregulation of fatty acid oxidation, ketone body production and the urea cycle.The SIRT3-mediated regulation of fasting metabolism is illustrated.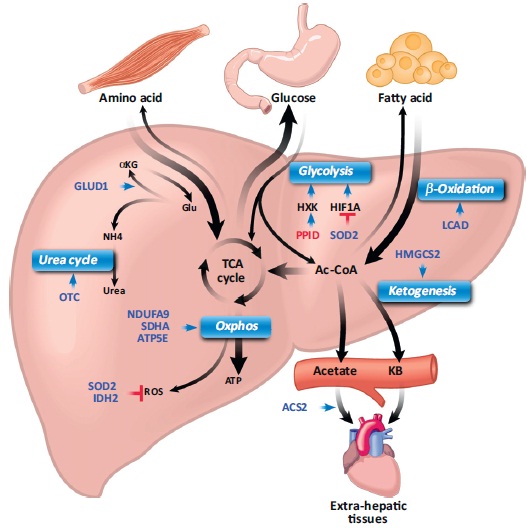
In times of abundance, glucose supplies bothacetyl-CoA to generate ATP, and carbon backbones to synthesize metabolic intermediates. In glucose-poor states such as fasting, ATP is instead generated predominantly from the b-oxidation of FAs derived from adipose tissue, and carbon backbones from the catabolism of amino acids from muscle. Acetyl-CoA is also converted to ketone bodies or, to a lesser extent acetate, which are then distributed to extrahepatic tissues through the bloodstream. SIRT3 regulates many of the enzymes involved in this switch to fasting metabolism. Proteins in blue, activated by SIRT3. In red, deactivated by SIRT3. More detailed descriptions of SIRT3 target genes are given in following Table.
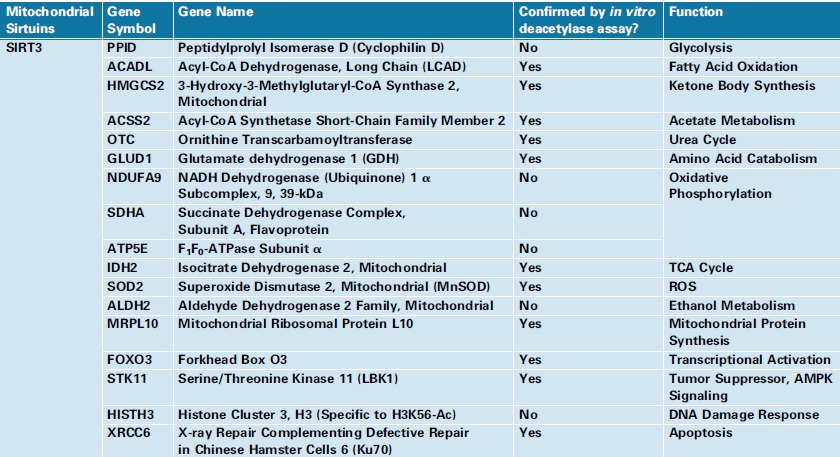
Therefore, SIRT3 is emerging as a key player in the metabolic adaptations to diet and lifestyle that may well influence mammalian lifespan.
SIRT6 has an essential role in postnatal life, and among the sirtuins its deficiency leads to the most dramatic phenotypes. SIRT6‑null mice are born with no visible abnormalities but soon after birth they develop a severe metabolic imbalance, hypoglycaemia and growth retardation, and the majority dies at approximately 1 month of age.
Enzymatically, SIRT6 acts as both a deacetylase and an ADP-ribosyltransferase, and a growing number of reports have highlighted roles for this sirtuin in DNA repair, telomere maintenance, genomic stability and cell senescence. SIRT6 attenuates nuclear factor-κB (NF-κB) signaling by interacting with its p65 (also known as RELA) subunit, and reducing RELA expression partially rescues the shortened lifespan of SIRT6‑deficient mice. In addition, SIRT6 co-represses hypoxia-inducible factor 1α (HIF1α) to suppress glucose uptake and glycolysis; the hypoglycaemia that limits lifespan in SIRT6‑null mice may be a result of unrestrained HIF1α expression.
However, the most impressive demonstration of a link between SIRT6 expression and longevity is a recent report showing that overexpression of SIRT6 extends the lifespan of male mice. In this study, SIRT6 overexpression lowered serum levels of insulin-like growth factor 1 (IGF1) and increased the expression of insulin-like growth factor-binding protein 1 in male mice, bringing the values closer to those observed in control female mice. By contrast, SIRT6 overexpression in female mice had no further effect on these parameters nor did it affect longevity. Attenuation of IGF1 signaling is associated with increased longevity in many animal models, suggesting that these effects could have a direct role in SIRT6‑induced lifespan extension.
Thus, SIRT6 has a major influence on mammalian physiology, is essential for normal lifespan and may directly influence longevity.References
Shi, T., Wang, F., Stieren, E. & Tong, Q. SIRT3, a mitochondrial sirtuin deacetylase, regulates mitochondrial function and thermogenesis in brown adipocytes. J. Biol. Chem. 280, 13560–13567 (2005). Sirt3 a mitochondrial situin_Shi_2005.pdfLiszt, G., Ford, E., Kurtev, M. & Guarente, L. Mouse Sir2 homolog SIRT6 is a nuclear ADP-ribosyltransferase. J. Biol. Chem. 280, 21313–21320 (2005).mouse sir2 homolog sirt6_Liszt_2005.pdf
Someya, S. et al. Sirt3 mediates reduction of oxidative damage and prevention of age-related hearing loss under caloric restriction. Cell 143, 802–812 (2010). This study establishes a role for SIRT3 in the protective effects of caloric restriction.
Sirt3 mediates reduction of oxidative damage_Someya_2010.pdf
Hirschey, M. D. et al. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature 464, 121–125 (2010).
Sirt3 regulates mitochondrial fatty acid_Hirschey_2010.pdf
Bellizzi, D. et al. A novel VNTR enhancer within the SIRT3 gene, a human homologue of SIR2, is associated with survival at oldest ages. Genomics 85, 258–263 (2005). A novel VNTR enhancer within the SIRT3 gene_bellizzi_2005.pdf
Rose, G. et al. Variability of the SIRT3 gene, human silent information regulator Sir2 homologue, and survivorship in the elderly. Exp. Gerontol. 38, 1065–1070 (2003). Variability of the SIRT3 gene_Rose_2003.pdf
Qiu, X., Brown, K., Hirschey, M. D., Verdin, E. & Chen, D. Calorie restriction reduces oxidative stress by SIRT3‑mediated SOD2 activation. Cell Metab. 12, 662–667 (2010). Calorie restriction reduces oxidative stress by sirt3_Qiu_2010.pdf
Hallows, W. C. et al. Sirt3 promotes the urea cycle and fatty acid oxidation during dietary restriction. Mol. Cell 41, 139–149 (2011).
Sirt3 Promotes the Urea Cycle and Fatty Acid Oxidation during Dietary Restriction_Hallows_2011.pdf
Palacios, O. M. et al. Diet and exercise signals regulate SIRT3 and activate AMPK and PGC‑1α in skeletal muscle. Aging 1, 771–783 (2009). Diet and exercise signals regulate SIRT3 and activate AMPK and PGC-1 alpha in skeletal muscle_Palacios_2009.pdf
Shimazu, T. et al. SIRT3 deacetylates mitochondrial 3‑hydroxy-3‑methylglutaryl CoA synthase 2 and regulates ketone body production. Cell Metab. 12, 654–661 (2010). SIRT3 Deacetylates Mitochondrial 3-Hydroxy-3-Methylglutaryl CoA _Shimazu T_2010.pdf
Mostoslavsky, R. et al. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell 124, 315–329 (2006). Genomic instability and aging-like phenotype in the absence of mammalian SIRT6_Mostoslavsky_2006.pdf
Kaidi, A., Weinert, B. T., Choudhary, C. & Jackson, S. P. Human SIRT6 promotes DNA end resection through CtIP deacetylation. Science 329, 1348–1353 (2010).Human SIRT6 Promotes DNA End Resection Through CtIP Deacetylation_Kaidi_2010.pdf
Mao, Z. et al. SIRT6 promotes DNA repair under stress by activating PARP1. Science 332, 1443–1446 (2011).
SIRT6 promotes dna repair under stress by activating parp1_Mao_2011.pdf
Michishita, E. et al. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature 452, 492–496 (2008).
SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin_Michishita_2008.pdf
Michishita, E. et al. Cell cycle-dependent deacetylation of telomeric histone H3 lysine K56 by human SIRT6. Cell Cycle 8, 2664–2666 (2009). Cell cycle-dependent deacetylation of telomeric histone H3 lysine K56 by human SIRT6_Michishita_2009.pdf
Yang, B., Zwaans, B. M., Eckersdorff, M. & Lombard, D. B. The sirtuin SIRT6 deacetylates H3 K56Ac in vivo to promote genomic stability. Cell Cycle 8, 2662–2663 (2009).The sirtuin SIRT6 deacetylates H3 K56Ac in vivo to promote genomic stability_Yang_2009.pdf
Kawahara, T. L. et al. SIRT6 links histone H3 lysine 9 deacetylation to NF‑κB‑dependent gene expression and organismal life span. Cell 136, 62–74 (2009). SIRT6 Links Histone H3 Lysine 9 Deacetylation to NF-kappa B-Dependent Gene Expression and Organismal Life Span_Kawahara_2009.pdf
Zhong, L. et al. The histone deacetylase Sirt6 regulates glucose homeostasis via Hif1α. Cell 140, 280–293 (2010).
The Histone Deacetylase Sirt6 Regulates Glucose Homeostasis via Hif1 alpha_Zhong_2010.pdf
Kanfi, Y. et al. The sirtuin SIRT6 regulates lifespan in male mice. Nature 22 Feb 2012 (doi:10.1038/ nature10815). This paper shows for the first time that overexpression of SIRT6 can extend lifespan in male mice. The sirtuin SIRT6 regulates lifespan in male mice_Kanfi_2012.pdf
Berryman, D. E., Christiansen, J. S., Johannsson, G., Thorner, M. O. & Kopchick, J. J. Role of the GH/IGF‑1 axis in lifespan and healthspan: lessons from animal models. Growth Horm. IGF Res. 18, 455–471 (2008).SIRT6 Links Histone H3 Lysine 9 Deacetylation to NF-kappa B-Dependent Gene Expression and Organismal Life Span_Kawahara_2009.pdf
Monday_12/31/2012
1) Provide the Km, Vmax, and Kcat values for SIRT3/NAD+.2) Literature search the Km, Vmax, and Kcat values for Sirtuins/NAD+, which will be used for discussion section.
3) Review the protocol of obtaining Ki for NAM and compare with reported data.
4) Perform a serial inhibition experiments to get Ki for the 6 reported (our JMB manuscript) inhibitor of SIRT3 under a range of concentrations.
- Write up a protocol for kinetic assay
- Check the availability of necessary reagents, if run short, should order them.
- Perform a titration experiment to select a reasonable concentration range of inhibitors.
RC (12-26):
XG, for the additions to the paper suggested below, we would like to know if you have recorded the Ki's for all 6 inhibitors that are shown in the current paper draft.
We would need the Ki's to compare to computation. It appears you have used a fixed concentration of inhibitor and measured the reduction in catalytic efficiency. It appears you have not obtained Ki's for these molecules and in order to do so, you would need to look at more concentrations of the inhibitors. Is this correct? If so, please list what experiments would need to be conducted here along with the approximate time required to do those experiments, and plan to do those when returning to work.
The plan for the next paper would be to record Ki's for a larger series of cogeneric inhibitors, with the new continuous enzyme-coupled assay (which I assume would give more accurate Ki estimates), and use these along with MM-GBSA calculations to predict Ki's for even more inhibitors. Do you have any other inhibitors in the lab?
One point we need to discuss: The Lineweaver-Burk relation below for inhibition appears to assume that the inhibitor can bind to the protein, but cannot engage in any other reactions. For NAM, we know that it participates
in the reverse reaction. Thus I would like to review how you get Ki for NAM based on the Lineweaver-Burk relation below; it would appear that NAM decreases v not only by competing with NAD+ for the binding pocket but also by increasing the rate of the reverse reaction. Did you report Ki for NAM? This has implications for extraction of Ki's for other inhibitors like iso-NAM that do not participate in the reverse reaction from the Lineweaver-Burk plots, because by displacing NAM, these molecules can decrease the rate of the reverse reaction and hence serve as activators. Unless the effect of these inhibitors in decreasing the rate of the reverse reaction is accounted for in the kinetic model, the Lineweaver-Burk relation would seem to predict that all such molecules that bind to the C pocket would decrease the reaction rate - whereas we know this is not true for iso-NAM in Sir2.
I think the problem originates in the fact that Michaelis-Menten kinetics does not explicitly account for the dissociation of P from the EP complex; i.e. it assumes ES -> E + P directly. Thus it appears Michaelis-Menten kinetics cannot describe how inhibitors displace P from E, which slows down the reverse reaction. However, this problem is not specific to sirtuins and NAM - it arises for any inhibitor that binds to the active site. I am thus assuming that most biochemists are ok ignoring this effect when using Lineweaver-Burk plots to estimate Ki's. On the other hand, I would like to know what other methods are used to estimate Ki's. In EK's computational papers, Ki's are reported for cogeneric series of inhibitors. Are these measured by directly assaying for binding affinity of the inhibitor to the protein, in the absence of substrate? If so, we might estimate Ki's using Lineweaver-Burk plots in this paper, but we should be aware that the correlation with MM-GBSA scores may be limited. In that case, we could work out another binding affinity assay for a follow-up paper, and use that experimental data to calibrate the MM-GBSA binding affinity calculations.
Based the info in hand, I think that in our future experimental work, determination of the reverse (was this referred to as the "base exchange" constant in XG's ppt?) rate constant for NAM for several sirtuins may be a key to activator design. One may be able to identify the sirtuins most susceptible to activation based on their values of this rate constant. Then, computational prediction of Ki's could be used to identify possible activators for screening with these sirtuins.
RC (12-18):
I've been looking over methods to compare the computationally predicted binding affinities to experimental data. Here are some of my thoughts so far:
1) We can use Km to estimate binding affinities for NAD+ only under the "rapid equilibrium" assumption - i.e., kcat/kr << 1. This is because:
Keq = 1/Km * (1 + kcat/kr)
since Km = (kr + kcat)/kf and Keq = kf/kr.
I made some edits to the document on Task List from Simulations regarding this.
The validity of the rapid equilibrium assumption is not clear in the case of Sir2/SIRT3 (typically only the so-called "quasi steady state approximation" holds). Any attempts to estimate binding affinities based on experimental data should be done for SIRT3, since the reaction mechanism involves direct binding to the AC pocket (i.e. we can assume there is only one ES complex, where E is enzyme plus peptide and ES is enzyme plus peptide plus NAD+).
XG, can you provide the Km and kcat values for SIRT3 / NAD+. I may use Km for such calculations and would like to have an idea of the order of magnitude of kcat. I didn't see these values in the paper. We may want to report them since they are the experimental values that are most important in comparison to computational binding affinities for NAD+.
To begin, we will compare the order of magnitude of the Delta G's obtained from the rapid equilibrium assumption to the Delta G's computed by Glidescore. Again, there is as of yet no justification for the rapid equilibrium assumption here.
2) If we can work out the issues with computational binding affinity predictions, we will be able to estimate the forward and backward rate constants for NAD+ binding. This is again because of the
relation between Keq, Km, kcat and kr above: Experimentally, we have Km and kcat, and computationally we will obtain Keq = exp(-Delta G/RT). Thus we can solve for kr, and from Keq = kf/kr we can
get kf as well. Thus we can get rates of binding without expensive MD simulations with the help of the experimental data. Again, this is under the assumption that binding is a single-step reaction. We can portray our efforts to obtain kf,kr for NAD+ binding as a first step towards obtaining rate constants for all steps of the sirtuin reaction mechanism, which is important for the design of activators.
3) With the experimental data alone, I believe we can compute the Delta G for binding of NAM and iso-NAM. This is because from the Lineweaver-Burk relation:
1/v = Km/Vmax * (1 + [I]/Ki)*1/[S] + 1/Vmax
Here I believe we have estimated Ki based on the experimental data, and though I have not looked at the derivation of the equation recently, I believe Ki is the dissociation constant for the inhibitor, i.e.
Ki = [E][I]/[EI]
where [E] is again the concentration of enzyme-peptide complex. So I believe we can obtain
Delta G_nam = -RT ln (1/Ki)
and similarly for iso-NAM. If so, we can compare experimentally determined Delta G's for NAM or isoNAM binding to the computationally predicted ones. These are also smaller molecules, so the computational results should be more reliable.
XG, please confirm that Ki here is the dissociation constant for the inhibitor, and provide the Ki's for SIRT3 and Sir2. Eric, if you haven't already done so in your slide presentation, please provide the Delta G's for NAM and iso-NAM.
4) I noticed that the velocities (v) reported in the paper are in units of AFU/min. Can we convert these to M/s (also important for the reporting of kcat)?
5) If we want to determine NAD+ binding affinities experimentally, XG, what experimental methods are available? Recall XG was looking into binding affinity assays when studying Thermofluor. The assays could be relevant for our future work in inhibitor design as well.
It would appear that this result is more relevant to our first paper than other auxiliary assays, but we do not necessarily need the numbers now. Would we need to use a catalytically inert NAD+ analog?
Monday_12/10/12
1) The cultures from pGEX-6P3-PncA will be inoculated under two different strategies according to the experience with 6His-MBP-PNCA. (12/10/12-12/12/12)(a) High growth temperature for higher cell density and short growth time
(b) Low growth temperature for lower cell density for longer time
(c) Over produce the above cell cultures and pellet them and store at -80oC for future use.
2) Prepare fresh lysate buffers and obtain whole protein from cell debris. (12/13/12-12/14/12)
Monday_12/03/12
Discuss with Eric about the final draft of our JMB paper on Tuesday (12/04/12). Concentrate on finalizing the manuscript to get ready for Dr Raj's review.(12/04/12-)Thursday_11/29/12
1) Eric and I will have skype to discuss the final draft of our JMB paper tomorrow (11/30/12).2) Low expression level of MBP-H6-PncA fusion protein might because of the formation of inclusion bodies (dense, insoluble aggregates that are failed folding intermediates). Have tried to lower the growth temperature to room temperature and adjust IPTG induction conditions by inducing at lower cell densities (A600=0.5) or growing the cells to a higher cell density (A600>1) for a shorter period of time
3) Testing pGEX-6P3-PncA system is on the way.
Monday_11/26/12
1) Reference search for the most updated research paper on Sirtuin/SIRT3 (11/29/12).2) Trouble shooting the low expression level of MBP-H6-PncA fusion protein and continue establishing the new expression system of pGEX-6P3-PncA.(11/27/12-)
3) Eric and I will have skype to discuss the final draft of our JMB paper on Thursday (11/29/12).
Monday_11/19/12
1) About IsoNAM- Eric: provide the binding affinity of IsoNAM-Sir2 and NAM-Sir2 (Eric has done the docking study); and try to calculate binding affinity of IsoNAM-SIRT3 and NAM-SIRT3 (It might be off since the starting crystal structure has been modified.)
- Xiangying: provide discussion if they are in agreement with published results; will combine with her experimental results to disucss the possible mechanism of IsoNAM in SIRT3.
2) I will update the manuscript when the remaining portions are fully finished. I have worked through the weekend at home and will try to finish my part no later than Wednesday (11/21/12).
3) The latest version Outline of JMB_112012.doc. (11/20/12)
Thursday_11/15/12
1) JMB has included papers in which the enzyme reaction/ mechanism was discussed. There are many more and the followings are just few examples- Inhibition of enzyme-catalysed reactions by excess substrate: A theoretical and monte carlo study of turning points in v(S) graphs Original Research Article Journal of Molecular Biology, Volume 169, Issue 2, 15 September 1983, Pages 597-617.William G. Bardsley, Francisco Solano-Muñoz, Andrew J. Wright, Philip B. McGinlay
- Drosophila Alcohol Dehydrogenase: Acetate–Enzyme Interactions and Novel Insights into the Effects of Electrostatics on Catalysis Original Research Article Journal of Molecular Biology, Volume 345, Issue 3, 21 January 2005, Pages 579-598.Jordi Benach, Jan-Olof Winberg, John-Sigurd Svendsen, Sílvia Atrian, Roser Gonzàlez-Duarte, Rudolf Ladenstein
- Crystal Structure of Bacillus sp. GL1 Xanthan Lyase Complexed with a Substrate: Insights into the Enzyme Reaction Mechanism Original Research Article Journal of Molecular Biology, Volume 350, Issue 5, 29 July 2005, Pages 974-986. Yukie Maruyama, Wataru Hashimoto, Bunzo Mikami, Kousaku Murata
RC: Have these been posted or will everything be posted together on Mon? The latest version I saw was dated 110512 below.
3) All references have been formated as JMB template.
4) Need more time to refine Figures and Results (to incorporate with simulation data, like isoNAM modulation), then some discussions.
Tuesday_11/13/12
1)Discussed with Eric on 11/12/12 and tasks are included in "Note 2012.11.12".2) Continue working on the remain tasks and try to finalize the manuscript by Friday (11/16/12)
Thusday_11/08/12
1) Edit all my experimental paper sections to make sure they flow smoothly2) Check if enzymatic reactions are sometimes discussed in JMB from a structural perspective
3) Discuss with Eric about the whole picture of the paper.
4) Discuss with Eric about all the docking studies involving isoNAM and decide if keep them in the first paper. If we keep them, I need to find way to connect them with my results plus some published data.
Monday_11/05/12
RC_1) (pg. 39-40) Any reason to include further details on iso-NAM activation based on “Mechanism of SIRT1 activation” document? There is already some discussion here. Add discussion on base exchange reaction rates only if splitting into two papersXG: Document "Mechanism of SIRT1 activation" includes the possible SIRT1 activating mechanisms by Resveratrol and Isonicotinamide. It covers borh kinetic experiments and crystallographic work done by six individual research groups (Denu, Nussinov, Sauve, Sinclair,Wolberger,and Zhang's ) since 2003. Base exchange reaction rate was used as a key parameter to emphasis the critical role Iso-NAM played.
In our first paper, we have found that (1) NAM competitively inhibits hSIRT3; (2) the addition of 50-500 uM of Iso-NAM slightly decreases the hSIRT3 inhibition in the presence of 200uM NAM. The fluorescence assay we used provides an overall deacetylation reaction rate and the enzyme-couple continuous assay closely monitor the amount of NAM in the reaction system, which possibly allows us to study iso-NAM in detail.
In current paper, I would like to report the result (2) without driven mechanism deeply since the paper focuses on inhibition effect. For our second paper, we can put more efforts to discuss the activation effects of two SIRT1 activators: resveratrol and isoNAM in terms of the different mechanisms they followed.
RC: Ok that's fine; does this mean we need to put all docking studies involving isoNAM in paper 2? Does it also mean that we should not discuss possible drug design strategies based on derepression in this paper?
XG: I will discuss with Eric about all the docking studies involving isoNAM and decide if keep them in the first paper. If we keep them, I have to find way to interpret them with my results plus some published data.
RC_2) Clearer indication of when the fluorescence vs continuous assays are being used – which plots correspond to which assay?
XG: Figure 1, 2, 3, and 4 are from fluorescence assay. Figure 5,6, and 7 are from continuous assay.
RC: Ok, I hope after the removals of the continuous assay the results sections still read ok.
XG: I am on the half way of editing the results section, may add new figure(s) based on the finished experimental data.
RC_3) Pg 39-40: abrupt transition between inhibition theory and iso-NAM discussion. Also, are we contrasting iso-NAM with allosteric activators like resveratrol here? Should be stated.
XG: Resveratrol is heterotropic substrate-driven allostery and isonicotinamide, a weak binding antagonist of nicotinamide could derepress Sir2 activity subject to nicotinamide inhibition. See update (Outline of Biochemistry_110512)Outline of Biochemistry_110512.docfor details.
RC 4) I noted in the abstract we referred to NAM binding in AB vs AC pockets. Isn't it more appropriate to say NAM binds in the B or C pocket?
XG: You are right. It will be more appropriate to say NAM binding in the B/C pocket. I will change them in Abstract.
RC 5) XG, please edit all your experimental paper sections to make sure they flow smoothly and there is no repetition and no unclear assumptions. Upon my first read
it seemed we did not read over the transitions between sections to make sure they all connect properly Thanks..
XG: I am editing all my experimental paper sections to make sure they flow smoothly
More comments of divided our work into two paper
I agree to put the continuous assay result into our second paper to not delay the submission.For the first paper, since it will be our first footprint in SIRTUIN community, I would like to make it strong. Other than continuous assay and some isoNAM work (see above), it will be nice to include most of the simulation work combined with experimental results. And we focus on inhibition.
Journals that fit for our topic in order
(1)Journal of Molecular Biology
(2) Biochemistry
(3)Journal of Bioloical Chemistry
(4) Journal of the American Chemical Society
RC: Ok, we all agree on this strategy. I also believe JMB is a good choice for the 1st journal since it is not uncommon to have detailed structural and sometimes computational analysis of proteins in JMB. I would just check to make sure enzymatic reactions are sometimes discussed there from a structural perspective.
XG: I will check if enzymatic reactions are sometimes discussed in JMB from a structural perspective
For our second paper, continuous assay results will be included. We can also put more efforts to discuss the activation effects of two SIRT1 activators: resveratrol and isoNAM in terms of the different mechanisms they followed. For this purpose, Eric might need to run more simulation.
Journals that fit for our topic in order
(1)Journal of Bioloical Chemistry
(2) Journal of the American Chemical Society
(3) Biochemistry
RC: Either1 or 2 are good choices. We will discuss this paper shortly. JACS would require more simulation.
XG: Just a very preliminary thought. If we decide to include isoNAM in our first paper, we will have Eric run many docking experiments for screening small molecular of our interest.
Thursday_11/01/12
Comments for revisions of Biochemistry paper
I have spent quiet a lot of time on Clone, Express and Purify PncA protein (nicotinamidase) for the continuous assay. It is one of the key reagents for our enzyme coupled continuous assay and it is not commercially available. By following the published protocol, I have successfully got pure PCR product from cDNA and cloned it into pAB vector. However, the vector from original reference is not available for us due to interest’s conflict between companies and universities. pAB vector has poor expression function of target protein. I have to modify new protocol for protein expression and purification, which took most of the time. For now, this simple clone has become itself an independent project and it will take longer time than what we expected. To not delay the submission, we can possible put it into another paper for this novel procedure.Therefore, we can split the paper as Dr Raj mentioned in Wiki. For the first paper, we will include fluorescence assay and docking study. The claims we are going to make in this paper:
1) NAM is a competitive inhibitor of hSIRT3 by experimental assay(Figure 1, 2, 3, and 4)
2) Docking results support our finding and …
3) By applying our kinetic study combined with docking, we show a success on testing out few small molecules for hSIRT3 inhibitor (as we mentioned in Figure 4_page 19, nicotinic acid n-oxide, pyridine n-oxide, and 1-methylnicotinamide).
4) Emphasis the significance of our method and future directions (little overview for our next JMB paper.)
For the second paper, we will put more words on both the development of enzyme coupled assay and MM-GBSA binding affinity calculation. The involvement of IsoNAM with nicotinamide (from Eric’s simulation) and with other small known inhibitors (Xiangying’s lab work using continuous assay_Figure 5, 6, 7 and more) can be highlight of the paper.
I am still working on continuous assay and get it done. At the mean time I will reconstruct my part based on our current version of manuscript if you agree to divide our current paper into two.
Monday_10/29/12
Experiment
(1) The 24 uninduced and 24 induced target protein pellet using different IPTG induction methods have been collected and waiting for next test.(10/29/12-)(2) The cell lysate methods have been developed and will test the efficiency at different aliqot of cell samples. (10/29/12-)
(3) Protein expression level will be evaluated from aformentioned 48 samples. (10/31/12-)
Monday_10/22/2012
Experiment
(1) The low expression level of MBP-H6-PncA fusion protein might due to (1)the effects of proteolytic degradation by the host; (2) the formation of inclusion bodies (dense, insoluble aggregates that are failed folding intermediates). Need to do (10/22/12-):- Lowering the growth temperature to between 20oC and 30oC
- Altering IPTG induction conditions (a) induction at lower cell densities (A600=0.5) usually results in greater yields of the fusion protein in a soluble form
(2) pGEX-6P3-PncA plasmid DNA was degisted and checked by argrous gel for correct size. Three out of 12 samples give strong signal. Those three samples (C4, E2, and E6) and parental pGEX6P3 (control) have transformed into E Coli BL 21 strain for protein expression.
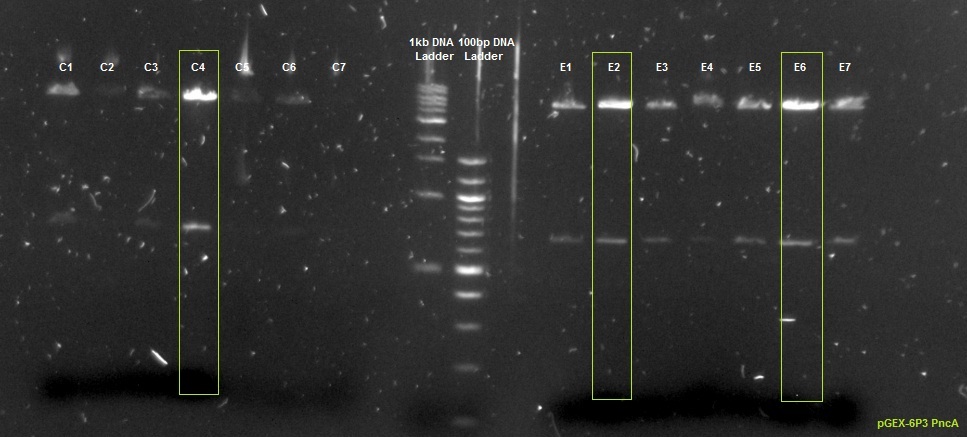
(3) Selected single colony was inoculated and IPTG induced expression will be performed. (10/20/12, 10/22/12)
(5) The expression level in new system will be evaluated this week. (10/22/12 -)
Thursday_10/18/12
Experiment
(1) MBP-H6-PncA protein with low expression level in BL 21 cell system has been extracted from culture and PncA protein will be purified after de-tag (H6). The efficiency of the affinity purification process will be evaluated. (10/19/12 -)(2) pGEX-6P3-PncA plasmid DNA has been purified from TOP10 system. The sizes of vector and target insert are correct! (10/17/12)
(3) The aforementioned ligation product has been transformed into BL 21 for protein expression. (10/18/12)
(4) Selected single colony will be inoculated and IPTG induced expression will performed. (10/19/12, 10/22/12)
(5) The expression level in new system will be evaluated next week. (10/22/12 -)
Monday_10/15/2012
Experiment
- (1)cell debris were removed by centrifugation. (10/15/12)
- Equilibrate HisTrap HP column with buffer A, run a linear gradient program for elution of MBP-H6-PncA protein. (10/16/12)
- MBP-H6-PncA containing fractions were pooled and H6-rTEV protease will be added for cleavage. (10/16/12)
- Dialysis with buffer at 4oC overnight (10/17/12)
- Tagless PncA protein resolved from above reaction using His-Trap HP column (10/18/12)
- (2) miniprep pGEX-6P3 - PncA plasmid DNA (10/16/12)
- Digest the purified plasmid with EcoR1 and Not1 at 37oC overnight. (10/17/12)
- Check the size on Agarous gel and transform into BL 21 (10/18/12)
Thursday_10/11/2012
Experiment
(1) scale up overexpression for purification- Culture samples from glycerol stock (10/09/12)
- IPTG induction of MBP fusion (10/10/12)
- Pellet and sonicate cells to get whole protein from cell lysates. (10/11/12-10/12/12)
- Vector pGEX-6P3 has been digested and gel purified for subclone. (10/09/12)
- Ligation of pGEX-6P3 with PNCA cDNA was performed overnight. (10/09/12)
- Transformation of aforementioned ligation product and self ligation (control) into TOP 10 cells and cell culture. (10/10/12)
- No self-ligation was detected, which means that CIP of vector plasmid was successful. (10/11/12)
- Inoculate the single colony (10/11/12) and miniprep when OD reach 0.6 (10/12/12). Measure concentration of purified Plasmid DNA (10/12/12).
Other
- Submitted the abstract to "Enzymatic Catalysis" under CALT Division for ACS national meeting (2013 spring).
Monday_10/08/2012
Experiment
- Protein level expression (10/08/12)
- Non-induced samples (C14_1,2,3,4; D6_1,2,3,4; E9_1,2,3; E12_1,2,3; F20_1,2,3; F21_1,2,3)
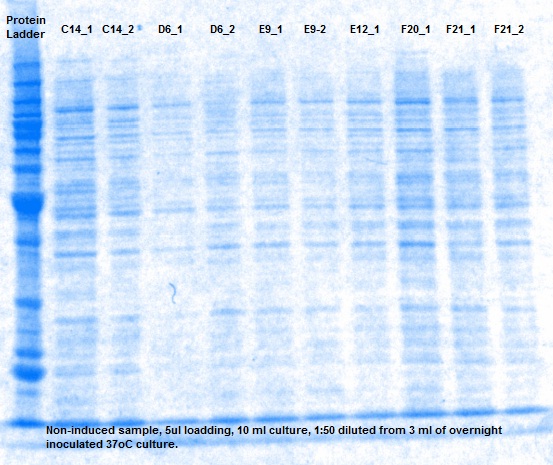

- Induced samples (C14_1,2,3,4; D6_1,2,3,4; E9_1,2,3; E12_1,2,3; F20_1,2,3; F21_1,2,3)
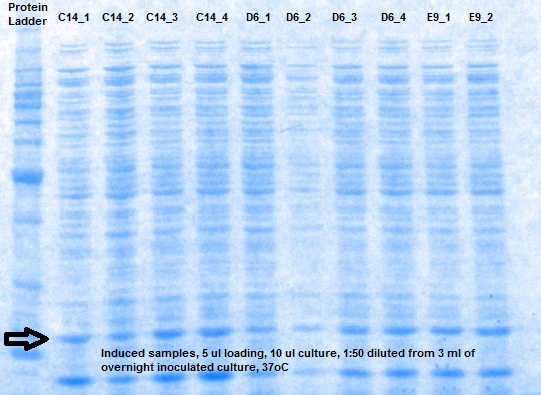
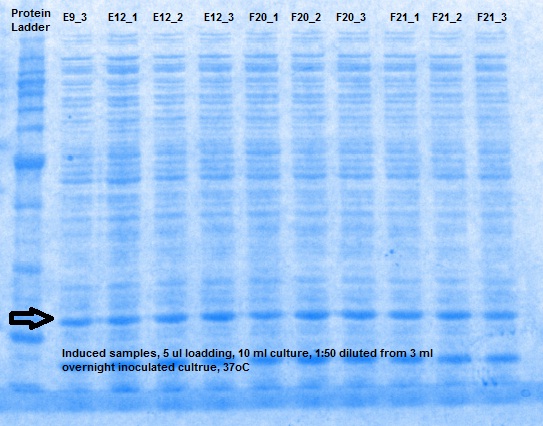
- Comparing with negative control (no induced samples), two significant bands were detected, which is close to the molecular weight of our target protein. However, the 6His-MBP-PNCA fusion protein (~67kDa) was not detected, which might due to non-specific protein degradation during the sample preparation. And the expression level is not high. (10/08/12)
- The expression system need to be optimized further. Will do
(2) Subclone the PNCA cDNA into more commonly used bacterial expression vector (pGEX-6P-3). (10/09/12-)
Thursday_10/04/2012
Experiment
- Still working on screening MBP fusion protein expression, with induced and non-induced samples, 20 of them has been done. Need more time to finish.(10/4/12-)
- Large-scale expression for affinity purification next week.
Manuscript
- Have discussed with Eric and updated in Task list from simulations. (10/3/12)
- Worked with Eric and got a draft of the abstract (10/4/12)Abstract_2013 ACS spring meeting.docx
Monday_10/01/2012
Experiment
- Screen the level of MBP fusion protein expression of 40 samples. (10/01/12-10/03/12)
- Purify proteins by affinity chromatography. (equlibrium column, setup gradient conditions...) (10/03/12-10/05/12)
- Enzyme activity measurement (10/05/12- )
Manuscript
- Will discuss with Eric to outline what new data/methods will need to be written up. (10/01/12)
- Have a draft of abstract ready for submission of 245th ACS meeting. (10/04/12)
Thursday_09/27/2012
Experiment
- 40 samples from 12 cultured plate were inoculated and overproduced. The production of MBP fusion protein has been induced by the addition of IPTG. (- 09/26/12)
- Pellets from 40 samples are stored in -20oC for further use. (09/27/12)
- Prepare buffer solution A and B, and extract MBP-His6-PNCA protein from cell lysate. (09/27/12)
- Prepare sample and check if the pNCA has been expressed by running SDS-PAGE gel. (09/28/12, 10/01/12)
- Equilibrium His-Tap cartridge and setup F(H)PLC system for protein purification next week. (09/28/12)
Monday_09/24/12
Experiment- Purified pNCA plasmide DNA was sucessfully transformed into E Coli BL 21 Strain. The positive colonies were detected on plate with respective antibiotics. (09/22/12) The plates were moved to 4oC.
- Cells from single drug-resistant colonies will be grown in LB broth supplemented with the appropriate antibiotics. at 30 oC/37oC over night. (09/24/12)
- The cells will be diluted 1:50 (30 oC) / 1:100 (37 oC) in the same medium and grown in shake flasks to early log phase (A600= 0.3-0.5) at 37oC .(09/25/12)
- Initiate the production of the MBP fusion protein (09/25/12)
- Prepare SDS-PAGE gel, sample preparation of uninduced control and induced samples. (09/26/12)
- Pellet the cells, sonicate, and extract MBP-His6-PNCA protein from cell lysate.(09/27/12).
Thursday_09/20/12
Experiment- Digested pNCA plamid DNA samples were checked on Gel for the right size (09/17/12)
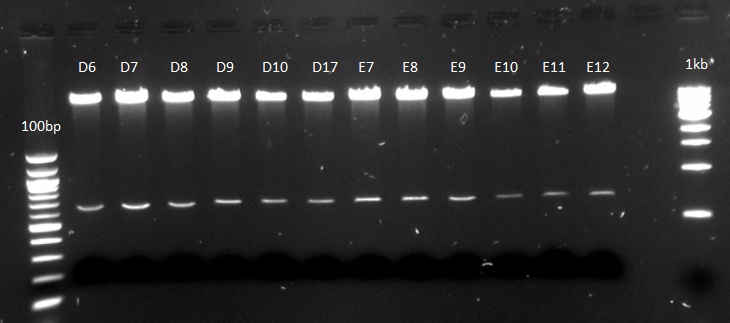
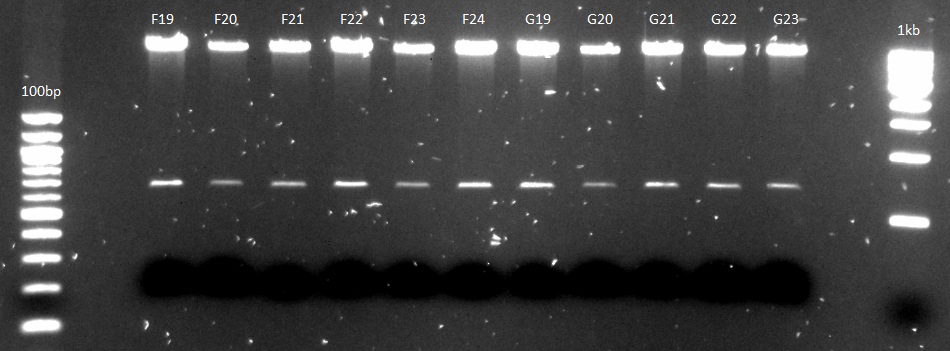
- Concentration of the above samples were measured. (09/18/12)
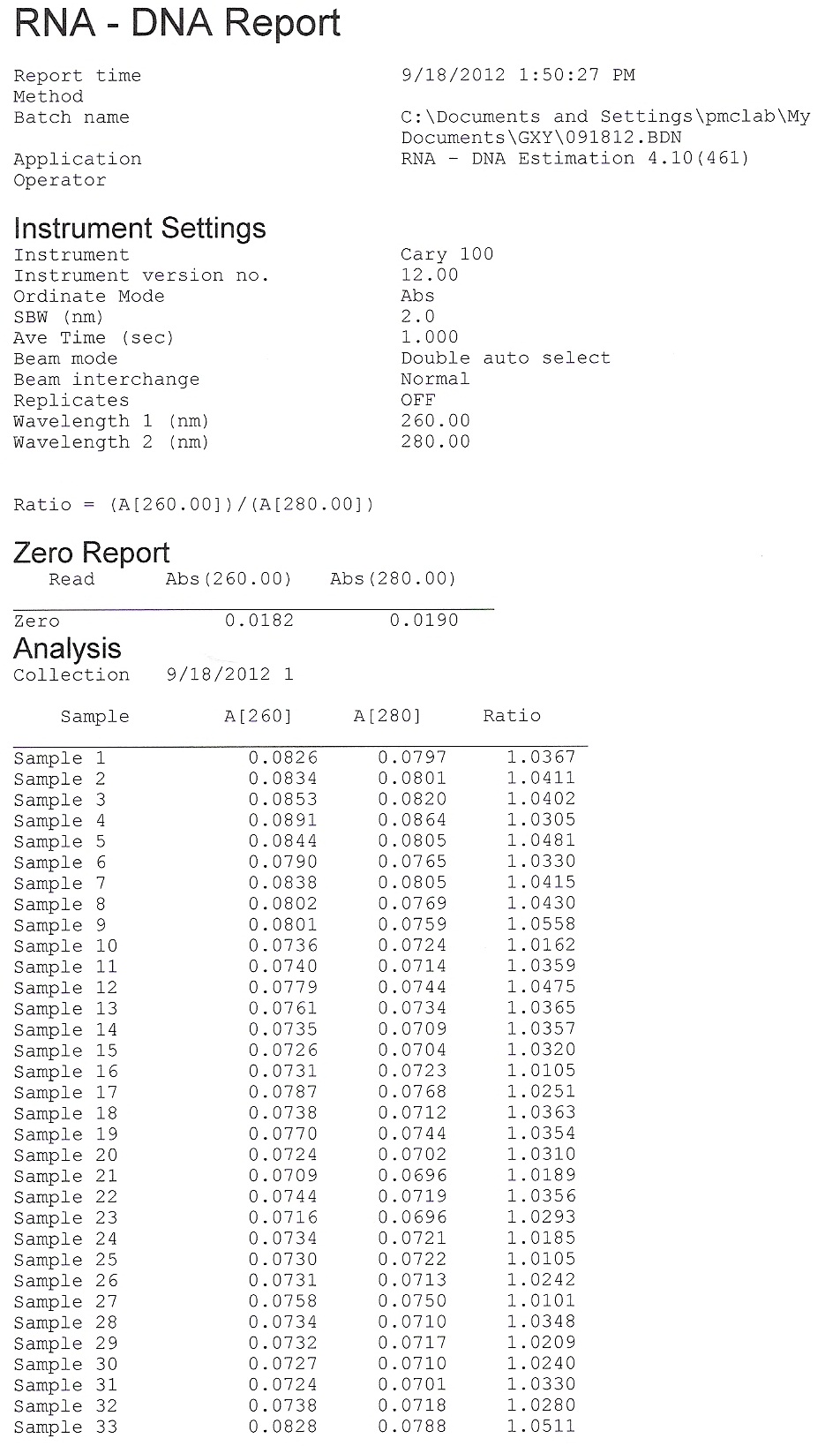
- From gel results, the selected aformentioned samples were sent out for sequencing. (09/18/12)
- Based on Seqencing results (09/20/12), the samples with correct sequence (C14, D6, E9, E12, F20, and F21) were tranfered into E Coli strain BL 21 cells.(09/20/12)
- Plate cell and grow for overnight. (09/21/12)
Manuscript
- Complications of Using Fluorescent labeled peptide substrate with insertion of figures. Complications of Using Fluorescent labeled peptide substrate.pdf
- Start to rearrange figures on macuscript.
RC:
XG, please provide your thoughts on whether we should aim to finalize the current version of the paper or wait for the results from these continuous time assays. Should we finish all the rest and leave room for this?XG:
I would like to put the continuous assay results into the manuscript since there's always an arguement of what role(s) the fluorophore play. Theoritically, the kinetic data from these two assays should be comparable. We can start to finalize the manuscript with ABSTRACT, INTRODUCTION, and MATERIAL/METHODS. RESULTS and DISCUSSION can be held for the data coming in because they highly depend on how data look like. I am on STEP 6 (see Monday09/10/12) and will try to get first set of data as soon as I can, then we can start finalize RESULTS and DISCUSSION at the time I repeat the experiments for the necessary statistic numbers.
Monday_09/17/12
Experiments
- Miniprep right product with positive colony and measure concentration of them.(09/15/12)
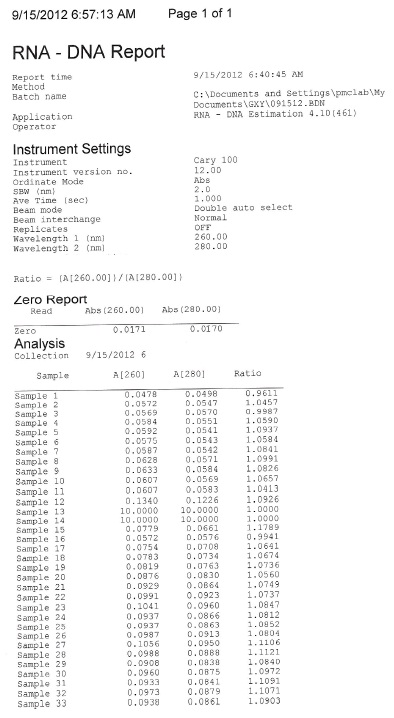
- Inoculate another batch of possible positive colonies _group #15, 19, 25, and 29. (09/17/12)
- Miniprep pNCA plasmid DNA (SampleD6, D7, D8, D9,D10, D17, E7,E8,E9,E10,E11,E12,F19,F20,F21,F22,F23,F24,G19,G20,G21,G22,G23) and digested with EcoR1 and Not1. (09/17/12)
- Gel confirmation of right sizes of both inserter and vector. (09/18/12)
- Over produce the culture and midiprep, concentration measurement, and gel confirmation. (09/19/12)
- Sequencing confirmation.(09/20/12)
Manuscript
- Complications of Using Fluorescent labeled peptide substrate (Figures will be added soon).
Back to 2005, both Denu and Kennedy labs (Borra, Smith et al, 2005; Kaeberlein, McDonagh et al, 2005) reported that it is necessary to use a peptide substrate with fluorephore to observe SIRT1 activation by resveratrol. Different assays were performed which were the Fluor de Lys assay, coumarin and rhodamine- based fluorescence assays, charcoal binding assay, and a HPLC-based assay.- It was found that SIRT1 activation was independent of the peptide sequence investigated (three p53 peptide substrates), but was dependent on the presence of a fluorescent label, thereby showing that resveratrol failed to activate the deacetylase activity of SIRT1 by using an unlabeled substrate. Substrate competition [[#|studies]] demonstrated that the attachment of a fluorophore decreased the binding affinity of the corresponding peptide, but in the presence of resveratrol, the tagged substrate bound more tightly (Borra, Smith et al. 2005).
- Borra et al. proposed a model for the effect of resveratrol on a coumarin labeled peptide. Without resveratrol, the coumarin attached to a p53 peptide would be solvent-exposed, and would exist in an energetically unfavorable conformation. Resveratrol may induce a conformational change that creates a binding pocket, which better accommodates coumarin, thereby resulting in enhanced substrate binding. Therefore, it is possible that a SIRT1 substrate containing the appropriate hydrophobic or aromatic amino acids might in fact be a target for resveratrol and other small-molecule-based activation (Borra, Smith et al, 2005).
- By using [3H]acetate and [14C]nicotinamide release assays, Kaeberlein et al. also demonstrated fluorescent tag-dependent activation of SIRT1 by resveratrol, and determined that the Km for the Fluor de Lys p53 substrate was approximately 8.5-fold higher than a p53 peptide lacking the fluorophore (Kaeberlein, McDonagh et al, 2005).
- More recently, Pacholec et al. published an independent study of the Sirtris compounds. By using an HPLC method to separate acetylated and deacetylated products, Pacholec et al. reported that like resveratrol, several Sirtris compounds led to SIRT1 activation in the presence of a peptide substrate covalently linked to a fluorophore, but not with an unlabeled peptide. NMR chemical shift studies (CH3) investigated the perturbation of the acetylated lysine group in peptide substrates with and without a covalently linked fluorophore. A resonance shift was detected when SRT1460 was incubated with a TAMRA-p53 peptide, but not with an unlabeled p53 peptide, thus providing evidence that SRT1460 interacted with the fluorophore.
- Surface plasmon resonance was used to demonstrate an interaction with the fluorophore, as concentration-dependent binding was observed with a TAMRA-containing peptide and not with a native peptide. ITC studies demonstrated that SRT1460 bound to SIRT1 in the presence of the fluorescently labeled peptide. However, SRT1460 did not bind to a SIRT1-unlabeled native p53 peptide complex; this indicates that SRT1460 bound only in the presence of the fluorophore (Pacholec, Bleasdale et al, 2010).
- Dai et al. confirmed the binding of SRT1460 and SRT1720 to TAMRA labeled peptides. However, the authors provide evidence that some compounds identified by high-throughput screens similar to Milne et al. (Milne, Lambert et al 2007) bind SIRT1 independent of the TAMRA tag, while others appear to increase SIRT1 activity in a substrate- dependent manner (Dai, Kustigian et al, 2010).
- Dai et al. also reported the substrate-dependent activation of SIRT1 with a few small-molecule compounds. These results suggest that SIRT1 activation could be dependent upon the hydrophobicity of residues located at the C-terminal of the acetylated lysine residue (Dai, Kustigian et al, 2010).
The fluorescence-based assays have been shown to be very efficient approaches for screening a large number of compounds. However, as with all highthroughput screening techniques, the results are best interpreted within the limiting context of the physical assay used. Numerous sirtuin assays have been reported, with each measuring different components of the reaction. To directly monitor deacetylation, MS, HPLC(Pacholec, Bleasdale et al. 2010; Borra and Denu, 2003) and microfluidic mobility shift assays (Liu, Gerber et al, 2008) have been described. A continuous microplate assay (Smith, Hallows 2009), [14C]nicotinamide release assay (Kaeberlein, McDonagh et al, 2005; McDonagh, Hixon et al. 2005), nicotinamide exchange assay (Landry, slama et al. 2000; Landry, Sutton et al. 2000), and TLC methods (Borra and Denu 2003; Landry, Sutton et al. 2000; Tanny and Moazed 2001) measure nicotinamide formation. To monitor the production of OAADPr, charcoal binding (Borra and Denu 2003), TLC (Landry, Sutton et al. 2000; Tanny and Moazed 2001), and HPLC assays can be used. These published assays afford a range of techniques capable of validating small-molecule effects.
References
M. T. Borra, J. M. Denu, Methods Enzymol. 2003, 376, 171.
M. T. Borra, B. C. Smith, J. M. Denu, J. Biol. Chem. 2005, 280, 17 187.
H. Dai, L. Kustigian, D. Carney, A. Case, T. Considine, B. P. Hubbard, R. B. Perni, T. V. Riera, B. Szczepankiewicz, G. P. Vlasuk, R. L. Stein, J. Biol. Chem. 2010; DOI: 10.1074/jbc.M110.133892.
M. Kaeberlein, T. McDonagh, B. Heltweg, J. Hixon, E. A. Westman, S. D. Caldwell, A. Napper, R. Curtis, P. S. DiStefano, S. Fields, A. Bedalov, B. K. Kennedy, J. Biol. Chem. 2005, 280, 17038.
J. Landry, J. T. Slama, R. Sternglanz, Biochem. Biophys. Res. Commun.2000, 278, 685.
J. Landry, A. Sutton, S. T. Tafrov, R. C. Heller, J. Stebbins, L. Pillus, R. Sternglanz, Proc. Natl. Acad. Sci. USA 2000, 97, Y. Liu, R. Gerber, J. Wu, T. Tsuruda, J. McCarter, Anal. Biochem. 2008, 378, 53.
T. McDonagh, J. Hixon, P. S. DiStefano, R. Curtis, A. D. Napper, Methods 2005, 36, 346.
J. C. Milne, P. D. Lambert, S. Schenk, D. P. Carney, J. J. Smith, D. J.Gagne, L. Jin, O. Boss, R. B. Perni, C. B. Vu, J. E. Bemis, R. Xie, J. S. Disch, P. Y. Ng, J. J. Nunes, A. V. Lynch, H. Yang, H. Galonek, K. Israelian, W. Choy, A. Iffland, S. Lavu, O. Medvedik, D. A. Sinclair, J. M. Olefsky, M. R. Jirousek, P. J. Elliott, C. H. Westphal, Nature 2007, 450, 712
M. Pacholec, J. E. Bleasdale, B. Chrunyk, D. Cunningham, D. Flynn, R. S. Garofalo, D. Griffith, M. Griffor, P. Loulakis, B. Pabst, X. Qiu, B. Stockman, V. Thanabal, A. Varghese, J. Ward, J. Withka, K. Ahn, J. Biol. Chem. 2010, 285, 8340
B. C. Smith, W. C. Hallows, J. M. Denu, Anal. Biochem. 2009, 394, 101.5807.
J. C. Tanny, D. Moazed, Proc. Natl. Acad. Sci. USA 2001, 98, 415.
Thursday_09/13/12
Experiments
- Select colonies from culture plates and inoculate in 33 groups. (09/11/12) _Done
- Miniprep and digested pNCA plasmid DNA (09/12/12)_Done
- Inoculate the positive colonies and miniprep pNCA plasmid DNA (09/13/13)
- Digested pNCA plasmid DNA (09/13/13)
- Gel confirmation for right product and over produce the right colonies in large scale.(09/14/12)
- Midiprep, purify pNCA plasmid DNA, concentration measurement, and cell transformation (09/15/12)
Manuscript
- Have discussed with Eric about the manuscript. The outline of the paper is posted by Eric on Task list from simulations.
- Have finished some of the insertion of relevant figures to cooperate with Discussion of Sirtuin activation mechanism. Will continue working on it. Mechanism of SIRT 1 activations_091312.docx
- Read and prepare a section for explaining the importance of having a continuing assay. (I have to find the time between experiments to sit down and do. Do not have a certain time frame and try best to finish it soon.)
Monday_09/10/12
RC: Can you provide an estimate of how much more time we will need to finish the experiments with the continuous time assay?XG: I started to clone, express, and purify pNCA enzyme for continuous assay end of June. Everything went well and extremly smooth till we got the pNCA plasmid DNA sequencing result. There was the mutation and I prodicted that it may caused by nonspecific error from Taq polymerase enzyme. That is Run 1. Then I changed another Taq enzyme and repeated the whole process. The same mutations were ditected from TOPO clone plasmid DNA sequencing. After communicated with ATCC (where we purchased the genomic DNA) tech support, it is possible the mutation came from genomic DNA (I am still waiting for the genomic DNA sequencing result to double confirm since it took 5-10 days). The Ligation efficiency was very poor on Run 2. I spent few extra days to figure out and make it work again. It will take another 3-5weeks to finish the assay and get some publishable data. Please see the following table for optimized time frame.
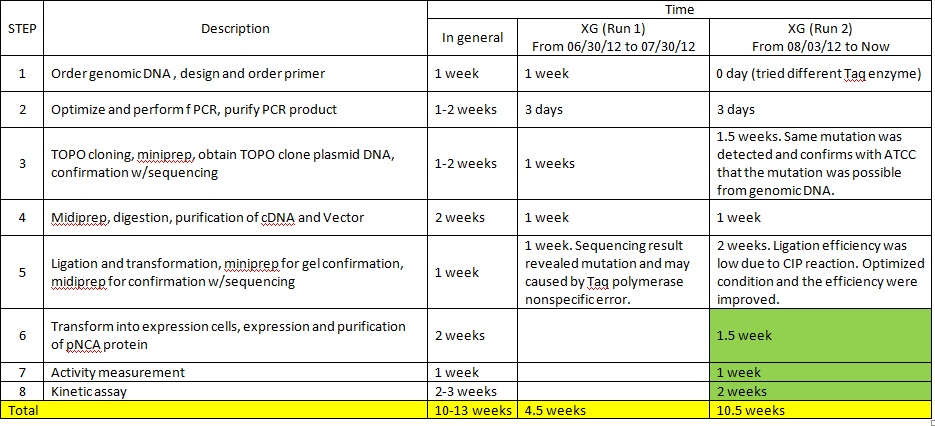
Experiments
- Digested plasmid DNA concentration was checked on Gel and on Spec.(09/08/12)
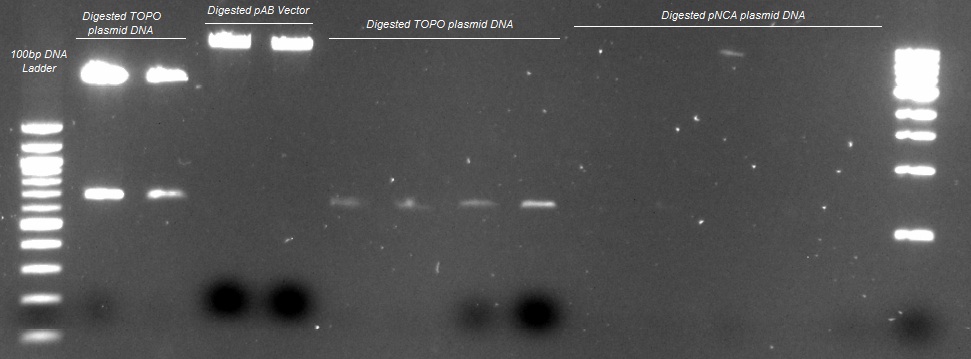
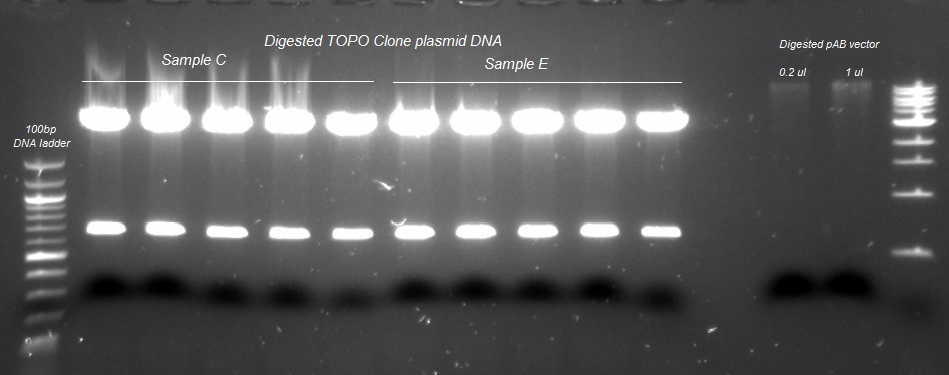
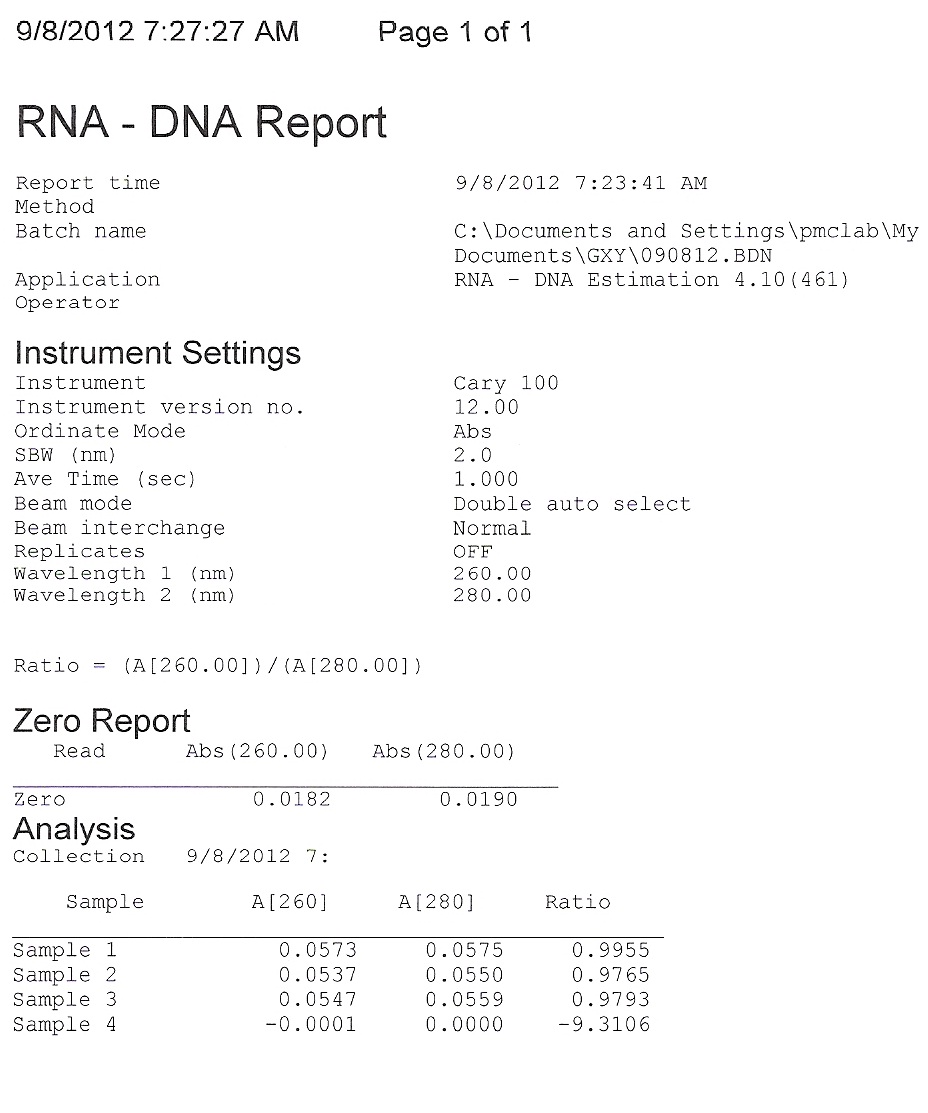
- Ligate inserter and vector overnight (09/08/12)
- Cell Transformation in Agar plate (09/09/12). Ligation efficiency was improved and need to diluted sample for single colony selection. (09/10/12)
- Check and inoculate positive colonies (09/11/12)
- Miniprep and digested pNCA plasmid DNA (09/11/12)
- Overproduce and Midiprep pNCA plasmid DNA (09/12/13)
- Gel and sequencing confirmation for right product (09/13/12 and 09/14/12).
Manuscript
- Setup skype with Eric to discuss the size of the paper and to coordinate efforts on refining/revising the existing manuscript sections. (09/10/12-09/13/12).
- Start inserting relevant figures to cooperate with Discussion of Sirtuin activation mechanism. (I have to find the time between experiments to sit down and do. Do not have a certain time frame and try best to finish it soon.)
- Read and prepare a section for explaining the importance of having a continuing assay. (I have to find the time between experiments to sit down and do. Do not have a certain time frame and try best to finish it soon.)
Thursday
RC: Can you provide an estimate of how much more time we will need to finish the experiments with the continuous time assay?Also, please have a discussion with Eric around Wed of this week to touch base on the size of the paper and to coordinate efforts on refining/revising the existing manuscript sections. We should be able
to start finalizing the content soon.
Thursday_09/06/12
Experiments
- The purified pNCA plasmid DNA was transferred into BL21 E Coli strain. As mentioned on 08/30/12, the culture grew very slow and only one colony was detected on the no diluted Agar cultrue plate (08/31/12)
- The plate was placed into 4oC over the weekend and put back in 37oC incubator for bigger colony size (09/04/12).
- That colony was inoculated into 3 ml LB broth but did not grow (09/05/12).
(b) The efficiency of transformation was low. Perform BL21 transformation again (09/05/12) with adjusted protocol.
(c) Digest Midiprep pNCA plasmid DNA (used for BL21 transformation ) overnight. (09/06/12)
(d) Check DNA concentration and digested product size. (09/07/12)
- Will come check the positive colony during the weekend. (09/08/12 and 09/09/12)
- Send Midiprep pNCA plasmid DNA out for sequencing (09/10/12)
Manuscript
- Start inserting relevant figures to cooperate with Discussion of Sirtuin activation mechanism. (09/07/12 ----)
- Read and prepare a section for explaining the importance of having a continuing assay. (09/07/12 ------)
Tuesday_09/04/12
Manuscript
Mechanism of Sirtuin activations: a comparison of resveratrol and isonicotinamideMechanism 1
In Sinclair’s 2003 nature paper, resveratrol was termed an allosteric effector of SIRT1 (as well as yeast Sir2). Previous results indicated that a conformational change in SIRT1 was induced by resveratrol binding, which stabilized fluorophore peptide binding (Borra, Smith et al. 2005).
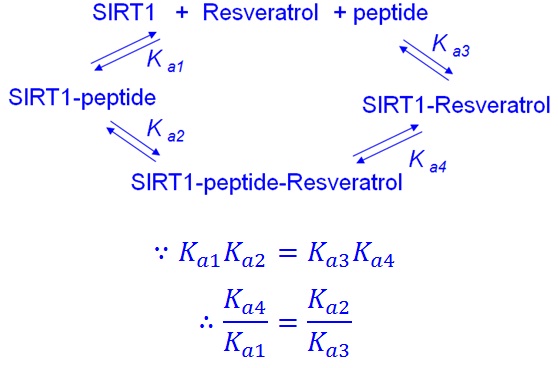
The enhanced binding of peptide to the activator-substrate complex is determined by the relative affinity of resveratrol for the peptide complex versus free SIRT1.
Mechanism 2
However, Miline et al. pointed out that resveratrol only bind to SIRT1 in the presence of fluorophore peptide (Milne, Lambert et al. 2007).
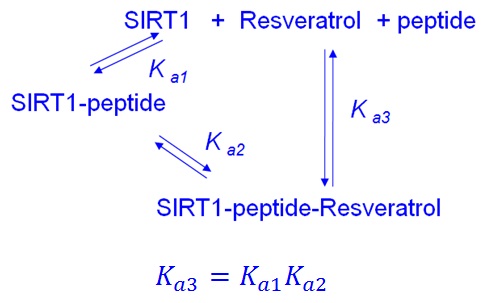
The net affinity for peptide binding to the resveratrol-bound complex is the multiplied association constants of peptide and resveratrol. Resveratrol that bind with similar affinity, but does not provide the same extent of activation (peptide-binding stabilization).
Conclusion: Since it is required to involve protein conformational change connecting two binding, the mechanism for the biochemical effect of resveratrol is heterotropic substrate-driven allostery. Accordingly, substrate binding opens a distinct site for activator-ligand binding, which stabilizes substrate binding by a thermodynamic component associated with substrate driven activator binding (Gunasekaran, Ma et al. 2004).
Sauve et al has identified the 3rd mechanism for activating Sir2 catalytic function: nicotinamide derepression, which is to interfere with nicotinamide inhibition of sirtuins. Isonicotinamide, a weak binding antagonist of nicotinamide could derepress Sir2 activity subject to nicotinamide inhibition (Sauve, Moir et al. 2005)
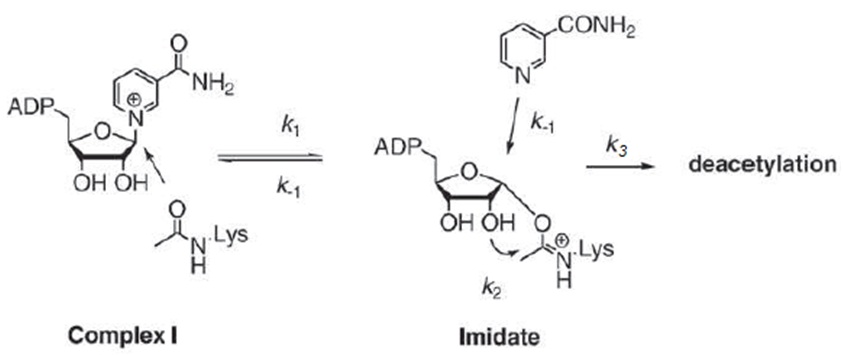
Studies indicate that in general the nicotinamide bond cleavage step is usually faster than the downstream chemistry of the imidate intermediate (Jackson, Schmidt et al, 2003; Sauve, Schramm, 2003; Borra, Langer et al, 2004). The nicotinamide bond cleavage step is not rate limiting and the nicotinamide base-exchange rate proceeds faster than the deacetylation reaction rate (Jackson, Schmidt et al, 2003; Sauve, Schramm, 2003). When nicotinamide base-exchange rate greatly exceeds the deacetylation rate (k-1 >k2), NAM inhibition is efficient. If the base-exchange rate is slower than the deacetylation rate (k−1<k2), inhibition of deacetylation by nicotinamide will not complete, such as the inhibition of Af2Sir2 deacetylase reaction by NAM does not exceed 50% (Sauve, Schramm, 2003). This type of inhibition is called hyperbolic or non-linear inhibition. This causes nicotinamide to be slower in reacting with the intermediate, relative to forward chemistry and results in hyperbolic inhibition.
Sauve and coworkers have suggested that Sir2/SIRT1 generally have very fast rate constants k1 and k−1 barriers relative to k2, pseudo-equilibration of the imidate and complex I would be predicted to occur when base exchange is at maximum rate. One possibility is that the imidate remains protonated during catalysis, and since an amide leaving group has negligible basicity even when compared to nicotinamide, the amide remains the superior leaving group and correspondingly the imidate is thermodynamically less stable.
- Wolberger laboratory has identified a residue which is likely important for interacting with the imidate proton, a Val 160 in the TmSir2 structure in which a backbone carbonyl is within hydrogen-bonding distance to the N of the imidate in the structurally determined thioimidate (Hawse, Hoff et al, 2008).
- Sauve group reported that nicotinamide is a poor inhibitor of Sir2Af2 and fails to inhibit deacetylation fully. The authors hypothesized that the intermediate is stabilized relative to the Michaelis complex in this case. There was no evidence that this stabilization is due to deprotonation of the imidate, since deacetylation remains active. They suppose instead that the intermediate is likely stabilized with respect to the complex I by some other means, which causes nicotinamide attack to be poorly competitive with 2 -OH attack on the imidate.
- One possibility has been suggested by structural work done by the Wolberger laboratory, who showed that a Phe residue interacts with the β-face of the ribose. This residue is highly conserved in sirtuins and can regulate the imidate reaction with nicotinamide. Mutation of this residue was found to profoundly accelerate the base exchange rate of the Sir2Tm enzyme, suggesting that this residue can shield the intermediate kinetically, and consequently produce stabilization. It is also possible that the Phe forms a π-cation interaction with the imidate which also contributes to stabilization, given that the sugar is calculated to retain an approximately 0.5 positive charge in the imidate complex (Hu, Wang 2008). Evidence that this Phe residue is mobile and adopts different geometries in different types of sirtuin complexes solved crystallographically fully supports the idea that this residue can modulate stability of imidate complexes. It has also been suggested that this Phe residue can prevent indiscriminate reactivity of the imidate with solvent, although increased hydrolysis relative to deacetylation in a Phe mutant has not been demonstrated.
References
1)Borra MT, Smith BC, Denu JM. Mechanism of human SIRT1 activation by resveratrol. J Biol Chem 2005; 280: 17187-95.
2) Gunasekaran K, Ma B, Nussinov R. Is allostery an intrinsic property of all dynamic proteins? Proteins 2004; 57: 433-43.
3) W.F. Hawse, K.G. Hoff, D.G. Fatkins, A. Daines, O.V. Zubkova, V.L. Schramm, W. Zheng, C. Wolberger, Structural insights into intermediate steps in the Sir2 deacetylation reaction, Structure 16 (2008) 1368–1377.
4) Howitz KT, Bitterman KJ, Cohen HY, Lamming DW, Lavu S, Wood JG et al. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature 2003; 425: 191-6.
5) P. Hu, S. Wang, Y. Zhang, Highly dissociative and concerted mechanism for the nicotinamide cleavage reaction in Sir2Tm enzyme suggested by ab initio QM/MM molecular dynamics simulations, J. Am. Chem. Soc. 130 (2008) 16721–16728.
6) M.D. Jackson, M.T. Schmidt, N.J. Oppenheimer, J.M. Denu, Mechanism of nicotinamide inhibition and trans-glycosidation by Sir2 histone/protein deacetylases, J. Biol. Chem. 278 (2003) 50985–50998
7) Milne JC, Lambert PD, Schenk S, Carney DP, Smith JJ, Gagne DJ, et al. Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes. Nature 2007; 450: 712-6.
8)A.A. Sauve, V.L. Schramm, Sir2 regulation by nicotinamide results from switching between base exchange and deacetylation chemistry, Biochemistry 42 (2003) 9249–9256.
Sauve AA, Moir RD, Schramm VL, Willis IM. Chemical activation of Sir2-dependent silencing by relief of nicotinamide inhibition. Mol Cell 2005; 17: 595-601.
Thursday_08/30/12
Experiments
- The mutations are from genomic DNA we purchased from ATCC. Have prepared the sample for Genom sequencing (08/30/12) to confirm and two choices
Have putified over produced pPNC1 plasmid DNA (08/27/12)
Transfered into E Coli BL 21 strain and cell culture. (08/29/12)
Slow grow and wait for collection of positive colony to inoculate overnight. (08/30/12)
Prepare solutions and buffers, get ready dialysis setup for protein purification (08/31/12)
Over produce the culture from 08/31/12 and Induce MBP-H6-PncA synthesis with IPTG overnight culture at 25oC. (09/04/12)
(B) Order Invitrigen kit for single point mutation to switch it back as it published on Pubmed.
It is a backup plan. Will do parallel to not slow down the whole process. (08/31/12 ---)
Note: Response from ATCC Tech is listed as following: The Bacteriology Collection Scientist took a look at your record and has said that we have no way of controlling or evaluating everything in GenBank. The sequences entered may contain ambiguous bases than an instrument or scientist had to make a judgment call to decide the sequence and that is not something that we would be able to investigate or know. We limit the passage number and stress on our cultures to avoid any mutations but we cannot check every gene for alignment with the published sequences.
[[#|Manuscript editing]]
- Inserted the equations and formated the section for different inhibition modes with kinetic equations for competitive, noncompetitive, and uncompetitive inhibition (In red and blue on page 33-35). (08/30/12)Outline of Biochemistry_083012.doc
- Redo the equation by equation editor in word. Use Eric's latest version as template without changing name. Outline of Biochemistry_083112.doc
Monday_08/27/12
Experiments
- Follow up with ATCC Tech for confirmation of PNCA genomic DNA Sequence. (08/30/12)
- Midiprep over porduced pPNC plasmid DNA and get ready for sequencing (08/27/12).
- Sequence the genomic DNA from ATCC.(08/27/12)
- Transfer purified pPNC1 into E.Coli BL 21 strain and cell culture overnight. (08/27/12)
- Collect the positive colonies and inoculate in 3ml of LB broth overnight.(08/28/12)
- Inoculate fresh LB medium and make 30% gerocel stock using the remainder of each overnight culture for long term storage (-80oC). (8/29/12)
- Induce MBP-H6-PncA synthesis and grown culture overnight. (08/29/12).
- Harvest cells and MBP-H6-PncA protein purification (08/30/12)
Manuscript editing
- The references in current format have been inserted into Biochemistry manuscript using EndNote. (08/27/12)Outline of Biochemistry paper_082712.doc
- Formate the section for different inhibition modes with kinetic equations for competitive, noncompetitive, and uncompetitive inhibition. (08/30/12)
Thursday_08/23/12
Experiments
- Double check the two samples C and E after sequencing.(8/23/12)TOPO clone purified digested prodcut sample C_E_082312.pdf
- Because the slow grow rate of ligations, the positive colonies were collected and inoculated in 3ml of LB broth.(08/23/12)
- Purify pPNC1 plasmid DNA and make 30% gerocel stock for long term storage (-80oC). (8/24/12)
- Over produce the pPNC1 plasmid DNA and collect pellets to store at -20oC. (08/27/12)
- Have sent the sequencing results over ATCC. They have microbiology specialist to get into it for their sequencing record of Genomic DNA we purchased. The record reference # is CA108579. (08/23/12). Will confirm with me if the mutations from their Geno DNA. (08/27/12)
Manuscript editing
- Rewrite "Protein expression and purification" in Method Section (page 6 and 7) marked in orange.(08/23/12)
- Modified Discussion section on page 37 marked in orange.(08/23/12)Outline of biochemistry paper.docx
- Start formating references needed for Biochemistry paper since we have EndNote installed at PMC-AT. (08/24/12)
- Find out if there is anyone who reported work of docking Iso-NAM into C pocket of Sir2. (08/27/12)
Monday_08/20/12
Experiments
- Purified pAB vector digested product was CIPed. The ligation was performed overnight. (08/16/12)
- Transfermation and cell culture on plate with respective antibiotic were finished. (08/17/12)
- The single colony grow very slow and another trasfermation will be needed for next step. (08/21/12)
- Collect positive clone and inoculate in LB broth. (08/22/12)
- Choice one, miniprep pPNCA plasmid DNA and send out for sequencing if the concentration is good. (08/23/12)
- Choice two, over produce The pPNCA plasmid DNA and midipred for sequencing. (08/27/12)
- Have contacted to ATCC tech support. Will send my sequencing results over there and PCR program with detailed reagent list for possible explaination. It is necessary to clairify that the mutation is from Genomic DNA they sale for publication propuse. (08/23/12)
Manuscript editing
- Comments on implications of our results for inhibitor and activator design (esp inhibitors – SIRT3 inhibitors cannot be designed based on AB pocket binding; for activators, expand current conclusion to indicate rational design of more effective activators than iso-NAM may be possible based on computational studies extending those presented – e.g., prediction of reaction rates for iso-NAM, NAM dissociation from C pocket). Contrast iso-NAM and resveratrol as Sirtuin activators (latter is allosteric, different design principles apply). (08/17/12)
- Updated into the Biochemistry paper manuscript with yellow highlight. The respective references are with yellow highlight. (Done on 08/17/12)
- Insert the pharma application section into Biochemistry manuscirpt with green color. The respective references are in green.(Done on 08/17/12)Outline of biochemistry paper.docx
- Update method section on page 6 (current position) where it marked as "Need to be more specifically addressed". (08/23/12)
- Update discussion section on page 36 where it marked as "Need more work here to state the novelty and significance of our work as a closing remark" (08/23/12)
- Start formating references needed for Biochemistry paper since we have EndNote installed at PMC-AT. (08/23/12)
Thurs_08/16/12
- Decide whether any add’l iso-NAM studies needed; not needed if the mode of inhibition through C pocket binding is already clear. Decide whether the SIRT3 iso-NAM activation data should be compared to that previously reported for Sir2. If C pocket binding, do we conclude leaving group kinetic properties are responsible for differences in activation? (08/16/12)
- XG: It was stated clearly in previously reported works that isonicotinamide can compete with nicotinamide for binding but cannot initiate the reverse reaction, thereby leading to apparent activation through relief of nicotinamide inhibition. Assuming that all Sirtuins are equally inhibited by nicotinamide, isonicotinamide would be a general Sirtuin “activator.” It is novel to include some simulation work to, at least on Sir2, provide structural basis evidences for the aforementioned statement. Looking back at Eric’s “Sirtuin Kinetics Paper draft 5-16-2012”, on page 4 “The implications iso-NAM activation kinetics for the design of activators of SIRT3”, few docking studies can be performed based on available co-crystallized Sir2 structure (1YC2_Sir2Af2 and 2H4F_Sir2Tm), which has not to my knowledge been developed anywhere else at this time. According to our current results, the addition of 50 -500 uM of isoNAM slightly decreases the inhibition effect in the presence of 200 uM NAM. At this point, if we could not firstly dock NAD+ into AB pocket of SIRT3 successfully, it will be hard to conclude leaving group kinetic properties are responsible for differences in activation on the case of hSIRT3. You can make the case on Sir2 though. Speaking of inhibition mode for iso-NAM on SIRT3, isoNAM IC50 is 13.8 mM, which indicates that isoNAM is a very weak (inefficient) inhibitor for SIRT3. It won’t do much by itself. It is not necessary to figure out experimentally whether iso-NAM is uncompetitive or competitive on NAM inhibition.
- Decide whether to cite/mention/differentiate our work form sirtris paper on mouse SIRT3 (08/16/12).
- XG: In Sirtris paper, the inhibition constants and mechanism of action for NAM and SRT1720 for mouse SIRT3 were determined. The results show that NAM is a competitive inhibitor of mouse SIRT3 respect to NAD+. Since the mouse SIRT3 has high sequence similarity with the C-terminus of human SIRT3, their result fall in good agreement with our finding, which is NAM competitively inhibits human SIRT3 deacetylation activity. I would like to point this out in our paper. No computational simulation work was reported in Sirtris paper. There is no need to describe the differences between our work from Sirtris paper.
- Start writing pharma applications section including description of applications of inhibitors and activators (08/16/12). Adapt this part into biochemistry paper template (08/17/12)
- Pharma application.docx
- The same mutation from sequencing results was found which indicated it might come from genomic DNA we purchased from ATCC. Have contacted to ATCC technical support and asked for the original sequencing from the sample. At the mean time, will carry on for next step.(08/16/12)
- CIP the digested pAB vector product and ligation. (08/16/12)
- Chemical transform and culture cell. (08/17/12)
- Comments on implications of our results for inhibitor and activator design (esp inhibitors – SIRT3 inhibitors cannot be designed based on AB pocket binding; for activators, expand current conclusion to indicate rational design of more effective activators than iso-NAM may be possible based on computational studies extending those presented – e.g., prediction of reaction rates for iso-NAM, NAM dissociation from C pocket). Contrast iso-NAM and resveratrol as Sirtuin activators (latter is allosteric, different design principles apply). (08/17/12)
Mon_08/13/12
- The purified digested TOPO clone plasmid DNA has been checked on the agarous gel for purity. The gel picture is posted on the "Lab" page.
- pAB vector has been digested and run on gel. The gel picture is posted on the "Lab" page.The respective bands have been cut and will be purified on Monday (08/13/12)
- A subsection in Discussion of the Biochemistry paper has been added with blue color (08/13/12). Outline of biochemistry paper.docx
- Provide the equations for calculationof Ki for competitive/noncompetitive inhibitors (08/13/12). Equation for calculation Ki for competitive and noncompetitive inhibition.pdf
- Align the sequencing result of TOPO clone plasmid DNA with PNCA and pick the right one(s) for ligation (08/16/12).
- Comments on implications of our results for inhibitor and activator design (esp inhibitors – SIRT3 inhibitors cannot be designed based on AB pocket binding; for activators, expand current conclusion to indicate rational design of more effective activators than iso-NAM may be possible based on computational studies extending those presented – e.g., prediction of reaction rates for iso-NAM, NAM dissociation from C pocket). Contrast iso-NAM and resveratrol as Sirtuin activators (latter is allosteric, different design principles apply). (08/16/12)
- Decide whether any add’l iso-NAM studies needed; not needed if the mode of inhibition through C pocket binding is already clear. Decide whether the SIRT3 iso-NAM activation data should be compared to that previously reported for Sir2. If C pocket binding, do we conclude leaving group kinetic properties are responsible for differences in activation? (08/16/12)
- Decide whether to cite/mention/differentiate our work form sirtris paper on mouse SIRT3 (08/16/12).
- Start writing pharma applications section including description of applications of inhibitors and activators (08/16/12).
Thurs_08/09/12
- The purified PCR products using new Taq (CloneTech) have been cloned into TOPO vector.
- The positive TOPO clone plasmid DNA has been purified.
- The size of digested product from TOPO clone plasmid DNA has been checked.The New Taq has better efficiency than SuperTaq used before.
- The gel figures are posted on "Lab" page.
- The pPNCA plasmid DNA from other positive clone on different plate has been purified and measured. Samples 1, and 2 is ready for Sequencing.
- The concentration of TOPO plamid DNA has been measured and is good enough to send out for sequencing.
- Prepare TOPO Clone plasmid DNA (Samples C,E, H, L, S, 1, 2) and send them out for Sequencing.
- Purify the digested TOPO clone plasmid DNA with right size (08/10/12).
- Check the purity by agarous gel (08/10/12)
- Digest pAB vector with EcoR1 and Not1 (08/10/12) and store at -20oC for gel purification and size checking on Monday (08/13/12)
Monday_08/06/12
- Post the latest version of the manuscript. (08/06/12)Outline of biochemistry paper.docx
Note: 1) The current manuscript has a big portion of experimental results. It depends on how much Eric's input to decide if we put into Biochemistry or stay with Journal of Biological Chemistry.
2) Not like Outline of JBC paper, I did not outline what/where Eric should put his part in details.
3) More results will added in when experiments are done.
4) Discussion need to be modified when Eric's results come in.
- Clone PCR product into TOPO vector (08/06/12)
- Transform TOPO clone into cell chemically and plate on Petri Dish with respective antibiotics (08/06/12)
- Midiprep the overproduced culture pellet from 08/03/12 and measure the concentration of DNA (08/07/12)
- Collect the positive clone and inoculate in LB broth (08/07/12)
- Miniprep and digest with EcoR1 and Not1 (08/08/12)
- Check the right size of digested product (08/09/12)
- Inoculate the cultue with right size of digested product (08/09/12)
Thurs_08/02/12
- The reagents for next step of experiments have beenordered from Invitrogen and Sigma.
- The program of PCR reaction using new Taq enzyme (CloneTech_ Advantage HF 2 polymerase) has beenmodified.
- The PCR reactions using different Taq enzymes have beenperformed.
- The purity of one set of PCR product was checked on agarous gel. The picture is posted on page of"Lab".
- Another set ofPCR product (different Taq enzyme with much longer reaction time) will be checked on Friday (08/03/12).
- Two set of PCR product will be purified together on Friday (08/03/12).
- The clones from other plates were incoulated and is overproduced in 50 mL culture overnight.
- Theoverproduced culture will be pelleted when OD reach ~0.6 tomorrow and Midiprep on Monday (08/06/12).
- PCR product will not be sent out for sequencing. Thesequencing will be held till PCR product is cloned into TOPO vector.
- Post the latest version of the manuscript based on those of another sirtuin researchers on Monday (08/06/12).
Mon_07/30/2012
Scheduled experiments
A single point mutation was detected from sequencing result (GTC vs GTT).(1) Sequencing the genom DNA_PNCA purchased from ATCC to check if the single point mutation is there already. If so, we will just carry on and test its activity.
(2) Check the Taq used for PCR and setup a new PCR reaction using other Taq enzyme available in the lab.
(3) Order reagents for clavage of 6His tag after separtion of target enzyme with others.
(4) Get ready for protein purification, like test HisTrap HP affinity column; prepare buffer solutions for washing, eluting.
Expected progress on Thurs
- Order reagents
- PCR reaction using new Taq enzyme (modify PCR program)
- Estimate the size ofPCR product by agarous gel
- Gel purify PCR product
- Measure concentration ofpurified PCR product
- Send aformentioned PCR product and genomic DNA out for sequencing
Questions/Comments
RC (8-10): XG paper-related tasks for Mon/Thurs (also read related comments under Simulation Tasks). Subdivide the tasks between Mon/Thurs sections of the task list above:
- Add a (sub)section describing the possible modes of inhibition of enzymes in some detail (this document was partly written before), so Eric’s computational section can refer to it
- Provide the equations for calculation of K_I’s for competitive/noncompetitive inhibitors (this document was written before)
- Start writing pharma applications section including description of applications of inhibitors and activators
- Comments on implications of our results for inhibitor and activator design (esp inhibitors – SIRT3 inhibitors cannot be designed based on AB pocket binding; for activators, expand current conclusion to indicate rational design of more effective activators than iso-NAM may be possible based on computational studies extending those presented – e.g., prediction of reaction rates for iso-NAM, NAM dissociation from C pocket). Contrast iso-NAM and resveratrol as Sirtuin activators (latter is allosteric, different design principles apply).
- Decide whether any add’l iso-NAM studies needed; not needed if the mode of inhibition through C pocket binding is already clear. Decide whether the SIRT3 iso-NAM activation data should be compared to that previously reported for Sir2. If C pocket binding, do we conclude leaving group kinetic properties are responsible for differences in activation?
- Decide whether to cite/mention/differentiate our work from Sirtris paper on mouse SIRT3. We have provided more mechanistic analysis through computation
Prepare sample and check if the pNCA has been expressed by running SDS-PAGE gel. (09/28/12, 10/01/12)
Prepare sample and check if the pNCA has been expressed by running SDS-PAGE gel. (09/28/12, 10/01/12)
![[carbonyl-14C]NAD_PE.jpg [carbonyl-14C]NAD_PE.jpg](files/%5Bcarbonyl-14C%5DNAD_PE.jpg)